Abstract
Sida szechuensis Matsuda is an economically and medicinally important plant. Here, we report the first chloroplast (cp) genome of the genus Sida (S. szechuensis). The complete cp genome is 159,878 bp in length with an overall GC content of 36.9% and consists of a large single copy region (LSC, 89,426 bp), a small single copy region (SSC, 114,715 bp), and a pair of inverted repeat regions (IRa and IRb, 25,288 bp). The genome encodes 111 unique genes, including 78 protein-coding genes, 29 tRNA genes, 4 rRNA genes, and 1 pseudogene. Phylogenetic analysis constructed using the maximum likelihood (ML) method showed that Sida was closely related to Malvastrum and Malva.
Sida szechuensis Matsuda 1918 belongs to the family Malvaceae, genus Sida. There are 100 genera in the Malvaceae, between 100 and 150 species in the Sida worldwide, and 14 species (six endemic) of Sida in China (Editorial Committee of Flora of China Citation2007). Sida szechuensis is a medicinal plant that grows in the highland approximately one to two thousand meters above sea level in Southwest China and is useful for treating ulcers, furuncle, dysentery, and blood stasis in ethnopharmacology (Xiao et al. Citation1997). Previous studies on S. szechuensis have focused on its chemical composition and medicinal value. However, analysis based on its complete cp genome is still lacking. In this study, the cp genome of S. szechuensis was obtained by next-generation sequencing technology. We analyzed the characteristics of the cp genome of S. szechuensis and revealed its phylogenetic relationships with other species in Malvaceae. These contents enrich the study of the cp genome information of the genus Sida and are important for further studies that evaluate the germplasm and molecular phylogeny of Malvaceae.
In this study, young leaf samples of S. szechuensis were collected from Dali, Yunnan Province, China (100°22′31.44″E, 25°45′5.45″N). The specimen was deposited at the Herbarium of Medicinal Plants and Crude Drugs of the College of Pharmacy, Dali University (De-Quan Zhang, [email protected]) under the voucher number ZSY110.
Total genomic DNA was extracted using the modified CTAB method from the dry and healthy leaves (Doyle Citation1987; Yang et al. Citation2014). Whole-genome sequencing was obtained via the Illumina HiSeq 2500 (Novogene, Tianjin, China) platform with the paired-end (2 × 300 bp) library. After removing the adapters and low-quality reads, the high-quality reads were assembled to the cp genome by GetOrganelle with the parameters 21, 45, 65, 85, and 105. (Jin et al. Citation2020). Then the assembled cp genome of S. szechuensis was annotated using Geneious 11.0.4 (Kearse et al. Citation2012) with the sequence of Malvastrum coromandelianum (MK860037) as the reference, and the stop codons of the ndhF, ccsA and ndhl genes were corrected manually. Finally, the cp genome sequences were deposited in GenBank (accession numbers, MT773597).
The cp genome of S. szechuensis has a total length of 159,878 bp and presents a typical quadripartite structure. It contains a pair of inverted repeat regions (IR, 25,288 bp) that were separated by a large single copy region (LSC, 89,426 bp) and a small single copy region (SSC, 114,715 bp). The cp genome comprised a total of 111 genes, including 78 protein-coding genes, 29 rRNA genes, 4 tRNA genes and 1 pseudogene (infA). The overall GC content of S. szechuensis was 36.9%, while those of the IR regions (42.9%) were higher than those of LSC (34.7%) and SSC (31.5%) regions. However, previous reports showed that the GC content varies in different regions of chloroplast genomes, but the IR regions have a high GC content due to the presence of rRNAs, which contain a high GC content (Mehmood, Abdullah Ubaid, Bao, et al. Citation2020; Mehmood, Abdullah Ubaid, Shahzadi, et al. Citation2020; Mehmood, Abdullah Shahzadi, et al. Citation2020).
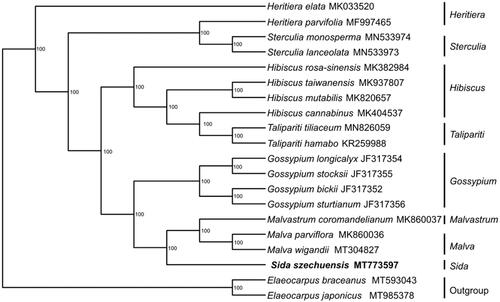
We downloaded 19 complete cp genome sequences from GenBank. Thirteen sequences of Malvaceae, four sequences of the genera Heritiera and Sterculia from the family Sterculiaceae and two sequences of Elaeocarpus braceanus and Elaeocarpus japonicus from the family Elaeocarpaceae were used as outgroups. All sequences were initially aligned using MAFFT V.7.149 (Katoh and Standley Citation2013). Then, the maximum likelihood tree was generated by RAxML (Stamatakis Citation2014) with 1000 bootstrap replicates under the GTRGAMMAI substitution model. The results of the phylogenetic analysis strongly supported that Sida is closely related to Malvastrum and Malva (). These results will provide valuable information for phylogenetic and evolutionary studies on Malvaceae.
Acknowledgement
We thank Dequan Zhang at Dali University for affording plant material for molecular experiments.
Disclosure statement
The authors are highly grateful for the published genome data in the public database. No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI under accession no. MT773597 (https://www.ncbi.nlm.nih.gov/nuccore/MT773597). The associated BioProject, SRA, and BioSample numbers are PRJNA746696, SRR15145519, and SAMN20236522, respectively.
Additional information
Funding
References
- Doyle J. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Editorial Committee of Flora of China. 2007. Flora of China. Vol. 12. Beijing: Science Press; p. 264–270.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CM, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Mark-Owitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Mehmood F, Abdullah Shahzadi I, Ahmed I, Waheed MT, Mirza B. 2020. Characterization of Withania somnifera chloroplast genome and its comparison with other selected species of Solanaceae. Genomics. 112(2):1522–1530.
- Mehmood F, Abdullah Ubaid Z, Bao Y, Poczai P, Mirza B. 2020. Comparative plastomics of Ashwagandha (Withania, Solanaceae) and identification of mutational hotspots for barcoding medicinal plants. Plants. 9(6):752.6.15.
- Mehmood F, Abdullah Ubaid Z, Shahzadi I, Ahmed I, Waheed MT, Poczai P, Mirza B. 2020. Plastid genomics of Nicotiana (Solanaceae): insights into molecular evolution, positive selection and the origin of the maternal genome of Aztec tobacco (Nicotiana rustica). PeerJ. 8:e9552.7.23.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Xiao X, Shu G, Xia W. 1997. A pharmacognostical study on the Chinese herbal drug“Badushan” (Sida szechuensis). Chin Tradit Herbal Drugs. 28(6):358–362.
- Yang JB, Li DZ, Li HT. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Resour. 14(5):1024–1031.