Abstract
The complete mitochondrial genome of the lizard, Teratoscincus przewalskii, which belongs to the family Sphaerodactylidae was determined based on Illumina data in this study. The result showed that the closed double-stranded circular mitogenome was 16,779 bp in total length (GenBank accession number: MW491837) with 44.07% GC. The complete mitochondrial genome consisted of 13 protein-coding genes, 22 transfer RNA genes, two ribosomal genes, and one noncoding control region. Phylogenetic analysis using mitochondrial genomes suggested that T. przewalskii was most closely related to its congener T. keyserlingii. This work provides valuable molecular information for further research on species identification and molecular evolution.
The Przesalski’s wonder gecko, Teratoscincus przewalskii, belonging to the subfamily Teratoscincinae (Squamata: Sphaerodactylidae), is known to be mainly distributed in Mongolia and Xinjiang, Gansu and Inner Mongolia in China (Gamble et al. Citation2007, Citation2011, Citation2012; Pyron et al. Citation2013; Nazarov et al. Citation2017). This species mostly lives in arid Gobi gravel sand, fixed dunes, semi-quicksand zones and Gobi Desert near reclaimed land. In this paper, we described the characteristics of the mitochondrial genome of T. przewalskii and discussed the phylogenetic relationships among Gekkota species, in order to provide a basis for further studies on interspecific taxonomy and phylogenetic relationships of these taxa.
Teratoscincus przewalskii was collected in Ejin Banner, Alxa League, Inner Mongolia, China in September 2020 (42.23 N, 101.31E) and was deposited in the laboratory of the College of Life Sciences and Technology of Inner Mongolia Normal University, Hohhot, China (http://bio.imnu.edu.cn/, Hui Yu, [email protected]). The muscular tissues were obtained and preserved in 95% ethanol. Total genomic DNA was extracted using the Qiagen Blood & Tissue Kit (QIAGEN, Hilden, Germany). Genomic DNA samples after testing qualified, with the method of mechanical interrupt (ultrasonic) DNA fragmentation, then end of fragmented DNA fragment purification, repair, and 3′ end and A sequencing, connection joints, and then selected fragment by agarose gel electrophoresis and sequencing library was formed by PCR amplification. After the library is built, the library quality inspection should be carried out. The qualified libraries were sequenced using Illumina Novaseq platform. The mitogenome was assembled by SPAdes v3.10.1 software (http://cab.spbu.ru/software/spades/) using Teratoscincus roborowskii (GenBank accession number: MW491837) as reference (Bankevich et al. Citation2012). The complete mitochondrial genome was annotated using Mitos2 (http://mitos2.bioinf.uni-leipzig.de), The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/) under the accession no. MW491837.
The complete mitochondrial genome of T. przewalskii was a circular molecule with 16,779 bp in total length (GenBank accession number: MW491837) and contained 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes, and one noncoding control region. The overall nucleotides composition was 30.90% A, 25.03% T, 13.78% G, and 30.29% C, which showed a bias toward A + T (55.93%). These were consistent with values found in other vertebrate species (Böhme et al. Citation2007; Li et al. Citation2016). Among the 13 PCGs, the common start codons (ATG and GTG) could be assigned as the start codon for most of PCGs and ND2, ND3 begin with ATA, only ND1 begins with ATC. The stop codons of the PCGs were TAA (ATP8, ATP6, ND4L, ND4, ND6), TAG (ND2, ND5, Cytb), TA (ND1, ND3), AGG (COI), T (COII, COIII). The size of the 22 tRNA genes ranged from 66 (tRNACys, tRNAVal) to 75 (tRNALeu) nucleotides. All tRNAs were foldable, with a typical clover structure, and their anticodon was exactly the same as the vertebrate tRNAs sequenced (Yan et al. Citation2009, Li et al. Citation2016, Yang et al. Citation2021). Two rRNA genes, 12S rRNA (948 bp) and 16S rRNA genes (1545 bp) were located between tRNAPhe and tRNALeu, separated by tRNAVal. The control region (1369 bp) was located between tRNAPro and tRNAPhe.
Based on the complete mitochondrial genome of T. przewalskii and other 22 species of Gekkota, a phylogenetic tree was constructed using Maximum-likelihood (ML) method on RAxML v8.2.10 software (https://cme.h-its.org/exelixis/software.html)with 1000 bootstrap replicates (Stamatakis Citation2014). The result showed that T. przewalskii was most closely related to its congener T. keyserlingii and rooted with the other Gekkonidae species (Han et al. Citation2004; Macey et al. Citation2005; Harris and Rato Citation2008; Nazarov et al. Citation2017) (). This mitochondrial genome provides valuable molecular information for further research on species identification and molecular evolution.
Figure 1. Phylogenetic position of T. przewalskii based on a comparison with the complete mitochondrial genome sequences of 22 other Gekkota species. The analysis was performed using RAxML v8.2.10 software. The accession number for each species is indicated after the scientific name.
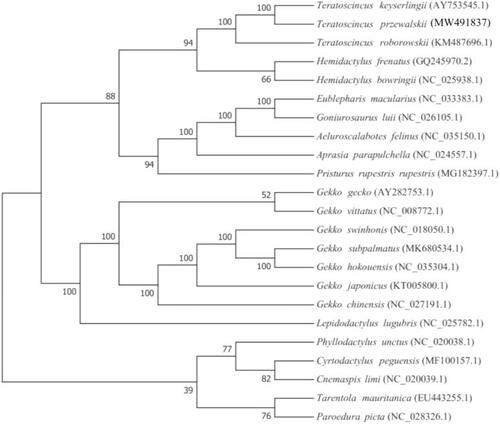
The inferred position of T. przewalskii herein was contradictory to previous and most recent publications (Macey et al. Citation1999, Citation2005; Tamar et al. Citation2021), which strongly support the sister relationship bewteen T. przewalskii and T. roborowskii. Whereas the previous publications used sequence lengths of NAD1 to COS1, we used the full sequence. This is the main reason for the difference in our analysis results. We still need more data in the future to verify the results.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MW491837.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Böhme MU, Fritzsch G, Tippmann A, Schlegel M, Berendonk TU. 2007. The complete mitochondrial genome of the Green Lizard Lacerta viridis viridis (Reptilia: Lacertidae) and its phylogenetic position within squamate reptiles. Gene. 394(1-2):69–77.
- Gamble T, Bauer AM, Colli GR, Greenbaum E, Jackman TR, Vitt LJ, Simons AM. 2011. Coming to America: multiple origins of New World geckos. J Evol Biol. 24(2):231–244.
- Gamble T, Bauer AM, Greenbaum E, Jackman TR. 2007. Evidence for Gondwanan vicariance in an ancient clade of gecko lizards. J Biogeography. 35(1):88–104.
- Gamble T, Greenbaum E, Jackman TR, Russell AP, Bauer AM. 2012. Repeated origin and loss of adhesive toepads in geckos. PLOS One. 7(6):e39429.
- Han D, Zhou K, Bauer AM. 2004. Phylogenetic relationships among gekkotan lizards inferred from C-mos nuclear DNA sequences and a new classification of the Gekkota. Biol J Linnean Soc. 83(3):353–368.
- Harris DJ, Rato C. 2008. Genetic variation within Saurodactylus and its phylogenetic relationships within the Gekkonoidea estimated from mitochondrial and nuclear DNA sequences. Amphib Reptilia. 29(1):25–34.
- Li HM, She Y, Hou LX, Zhang Y, Guo DN, Qin XM. 2016. The complete mitochondrial genome of Teratoscincus roborowskii (Squamata: Gekkonidae). Mitochondrial DNA Part A. 27(3):1916–1917.
- Macey JR, Fong JJ, Kuehl JV, Shafiei S, Ananjeva NB, Papenfuss TJ, Boore JL. 2005. The complete mitochondrial genome of a gecko and the phylogenetic position of the Middle Eastern Teratoscincus keyserlingii. Mol Phylogenet Evol. 36(1):188–193.
- Macey JR, Wang Y, Ananjeva NB, Larson A, Papenfuss TJ. 1999. Vicariant patterns of fragmentation among gekkonid lizards of the genus Teratoscincus produced by the Indian collision: a molecular phylogenetic perspective and an area cladogram for Central Asia. Mol Phylogenet Evol. 12(3):320–332.
- Nazarov RA, Radjabizadeh M, Poyarkov, Jr NA, Ananjeva NB, Melnikov DA, Rastegar-Pouyani E. 2017. A new species of frog-eyed gecko, genus Teratoscincus strauch, 1863 (Squamata: Sauria: Sphaerodactylidae), from central Iran. Russ J Herpetol. 24(4):291.
- Pyron RA, Burbrink FT, Wiens JJ. 2013. A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Evol Biol. 13:93.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tamar K, Els J, Kornilios P, Soorae P, Tarroso P, Thanou E, Pereira J, Shah JN, Elhassan EEM, Aguhob JC, et al. 2021. The demise of a wonder: evolutionary history and conservation assessments of the wonder gecko Teratoscincus keyserlingii (Gekkota, Sphaerodactylidae) in Arabia. PLOS One. 16(1):e0244150.
- Yan J, Zhou JL, Tian C, Zhou KY. 2009. Complete nucleotide sequence and gene organization of the mitochondrial genome of common house Gecko, Hemidactylus Frenatus. J Nanjing Norm Univ Nat Sci Ed. 32:77–82.
- Yang X, Lian Y, Chen M, Li X, Yu D. 2021. Characterization and phylogenetic analysis of the complete mitochondrial genome of sun loach (Yasuhikotakia eos). Mitochondrial DNA Part B. 6(1):13–14.
