Abstract
Ranunculus japonicus is an important medicinal herb widely used in East Asia. In this study, we report the first complete chloroplast genome sequence of Ranunculus japonicus using next-generation sequencing technology. The chloroplast genome size of R. japonicus was 156,981 bp. A total of 129 genes were included, consisting 84 protein-coding genes, eight rRNA genes, and 37 tRNA genes. Thirteen protein-coding genes had intron (ycf3 gene, rps12 gene, rps12 gene, clpP gene contained two introns). A further phylogenomic analysis of Ranunculaceae, including 10 taxa, was conducted for assessing the placement of R. japonicus. It will provide valuable genetic information for this medicinally important species.
Ranunculus japonicus Thunberg 1794 is a topical herb widely distributed in East Asia (Cao et al. Citation1992). It inhabits the moist grasslands and mountains. As a traditional Chinese medicine, it is widely used to treat various diseases, including malaria, jaundice, migraines and stomachaches for over 1800 years (Rui et al. 2010). In Korea, the young stems of R. japonicus are eaten as a vegetable (Yun et al. Citation2021). In addition, recent study showed that 27 validated compounds of R. japonicus were predicted to exhibit therapeutic effects against rheumatoid arthritis, which offered a potential clinical application (Wang et al. Citation2021). However, there are few relevant reports about the genetic information and phylogenetic relationships of R. japonicus and its relatives. In the study, the first complete chloroplast genome of R. japonicus was assembled and characterized. It will provide potential genetic resources for further population genetic study of this medicinally important species.
The R. japonicus individual was collected from Xuancheng, Anhuhi, China (GPS: 118°41′30.73″, N 30°34′15.49″). DNA was extracted from its silica dried leaves using DNA Plantzol Reagent (Invitrogen, Carlsbad, CA, USA) in accordance with the manufacturer’s instructions. The specimen and extracted DNA was deposited at Zhejiang Province Key Laboratory of Plant Secondary Metabolism and Regulation, Zhejiang Sci-Tech University (http://sky.zstu.edu.cn) under the voucher number ZSTU00034 (collected by Zhe-Chen Qi and [email protected]). Sequencing libraries were prepared using Illumina’s TruSeq Nano DNA Library preparation kit (350 bp median insert) following the manufacturer’s protocol. The plastome sequences were generated using the Illumina HiSeq 2500 platform (Illumina Inc., San Diego, CA, USA). In total, about 14.7 million high-quality clean reads (150 bp PE read length) were generated with adaptors trimmed. These clean data were de novo assembled to complete chloroplast genome using GetOrganelle (Jin et al. Citation2020). Geneious v11.1.5 (Biomatters Ltd, Auckland, New Zealand) was used to annotate the genome with R. sceleratus plastome (GenBank: MK253452.1) as a reference.
The full length of the complete R. japonicus chloroplast sequence (GenBank Accession No. MZ169045) is 156,981 bp, consisting of a large single copy region (LSC with 85,454 bp), a small single copy region (SSC with 18,897 bp), and two inverted repeat regions (IR with 26,315 bp). The overall GC content of R. japonicus chloroplast genome was 37.7%. A total of 129 genes were included in the genome (84 protein-coding genes, 8 rRNA genes, and 37 tRNA genes). Seventeen genes had two copies, which were comprised of 6 PCG genes (ndhB, rps7, rps12, ycf2, rpl2, rpl23), 7 tRNA genes (trnl-CAU, trnl-CAA, trnv-GAC, trnl-GAU, trna-UGC, trnR-ACG, trnN-GUU), and all 4 rRNA species (rrn16, rrn23, rrn4.5, rrn5). In the genome, nine protein-coding genes (rps16, atpF, rpoC1, petB, petD, rpl16, rpl2, ndhB, ndhA) had one intron, and ycf3 gene, rps12 gene, rps12 gene, clpP gene contained two introns.
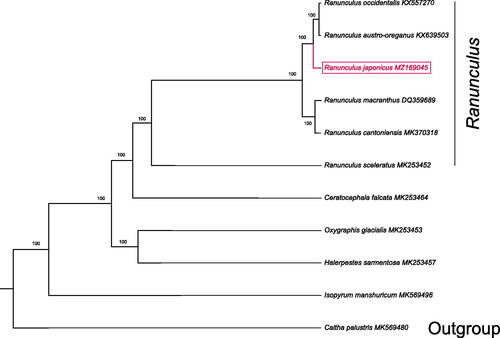
Ten species with available chloroplast genomes were selected to study the phylogenetic placement of R. japonicus in Ranunculaceae. Caltha palustris was used as outgroup for constructing the phylogenetic tree. Alignment of plastomes was generated by MAFFT v7.475 (Katoh and Standley 2013). A maximum-likelihood tree was generated by IQTREE v1.6.8 (Nguyen et al. Citation2015), with the best selected TVM + F+R2 model and 5000 bootstrap replicates. The result showed that R. japonicus was closely related to a clade formed by R. occidentalis, and R. austro-oreganus according the current sampling extent ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The associated BioProject, SRA, and Bio-Sample numbers are SUB9615995, PRJNA729222, and SAMN19114502 respectively. The DNA matrix and phylogenetic tree that support the findings of this study are openly available in figshare at https://doi.org/10.6084/m9.figshare.16585595.v1.
Additional information
Funding
References
- Cao BJ, Meng QY, Ji N. 1992. Analgesic and anti-inflammatory effects of Ranunculus japonicus extract. Planta Med. 58(6):496–498.
- Jin JJ, Yu WB, Yang JB, Song Y, Claude W, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Rui W, Chen H, Tan Y, Zhong Y, Feng Y. 2010. Rapid analysis of the main components of the total glycosides of Ranunculus japonicus by UPLC/Q-TOF-MS. Nat Prod Commun. 5(5):783–788.
- Wang ZY, Chu FH, Gu NN, Wang Y, Feng D, Zhao X, Meng XD, Zhang WT, Li CF, Chen Y, et al. 2021. Integrated strategy of LC-MS and network pharmacology for predicting active constituents and pharmacological mechanisms of Ranunculus japonicus Thunb. for treating rheumatoid arthritis. J Ethnopharmacol. 271:113818.
- Yun HS, Dinzouna-Boutamba SD, Lee S, Moon Z, Kwak D, Rhee MH, Chung DI, Hong Y, Goo YK. 2021. Antimalarial effect of the total glycosides of the medicinal plant, Ranunculus japonicus. Pathogens. 10(5):532.