Abstract
Lonicera similis Hemsl. belongs to the Caprifoliaceae family and used as a substitute for ‘jin yin hua’. Recent years, it demonstrates great economic value because of its rich chemical composition. However, the phylogenetic relationship between L. similis and other family members remains unclear. In this paper, we assembled the cp genome of L. similis using the high-throughput Illumina pair-end sequencing data. The circular cp genome was 155,207 bp in size, including a large single-copy (LSC) region of 88994 bp and a small single-copy (SSC) region of 18,633 bp, which were separated by two inverted repeat (IR) regions (23,790 bp each). A total of 121 genes were predicted, including eight ribosomal RNAs (rRNAs), 36 transfer RNAs (tRNAs), and 77 protein-coding genes (PCGs). In addition, the result of phylogenetic analysis indicated that L. similis formed a close relationship from another congeneric species (Lonicera confusa). This study provides helpful information for future genetic study of L. similis.
Lonicera japonica Thunb is a kind of well-known traditional Chinese medicine and has a common known name as ‘jin yin hua’ in Chinese. It has been used to treat fever, eliminating inflammation and antibacterial (Weng et al. Citation2011; Hu et al. Citation2012; Zeng et al. Citation2020). Lonicera similis Hemsl. is mainly founded in south shaanxi, south gansu and other regions in China, which belongs to the Caprifoliaceae family and has the same function with Lonicerae Japonicae Flos in modern pharmacological studies (National Pharmacopeia Committee 2015). Therefore, it is used as a substitute for ‘jin yin hua’ in mountainous areas of southwest China (Zhang et al. 2018). At present, the research on chemical composition has been in-depth, involving organic acid, flavonoids, saponins, volatile oil and so on (Lv et al. Citation2012). Among them, chlorogenic acid is the most important secondary metabolites of medicinal materials of honeysuckle (Cai et al. Citation2020) and its content in the L. similis has been proved higher than the current pharmacopeia standards(Ma et al. Citation2007). Although L. similis contains lots of valuable information and economic benefits through chemical composition research, there has rarely been any report for a genetic study of the plant. Therefore, the complete chloroplast (cp) with many advantages has become our ideal tool for phylogeny research (Dong et al. Citation2014; Saina et al. Citation2018). In this paper, the cp genome of L.similis Hemsl. has been assembled and determined. The study will be a helpful resource for future genetic research and determining phylogenetic relationships.
Healthy and fresh leaf samples were picked from Dangwu Town, Huaxi District, Guiyang City, Guizhou Province, China (26°23′34.49″N, 106°35′56.89″E, 1,158 m above sea level). The plants were identified using a species identification bench mark set up by Dr. Chunyan Han, Kunming Caizhi Biotechnology Co. Ltd, Kunming, Yunnan, China. A voucher specimen was deposited at a local herbarium of School of Life Sciences, Guizhou Normal University (contact person named Wenqing He and his e-mail is [email protected]) with accession numbers GZNUYZW202101002. The total genomic DNA (No. YX20210115901) was extracted using E.Z.N.A Plant DNA kit (FEIYANG, Guangzhou, China) and stored in the biochemical laboratory (room number: 1403) of School of Life Science, Guizhou Normal University. A total amount of 1000 ng DNA per sample was used as input material for the DNA sample preparations. Sequencing libraries were generated using NEB NextV RUltra DNA Library Prep Kit for IlluminaV R (NEB, Ipswich, MA). Total DNA was used to generate libraries with an average insert size of 400 bp. The library preparations were sequenced on an Illumina platform and 150 bp paired-end reads were generated.The program GetOrganelle will be used to assemble the filtered reads with Lonicera japonica as the initial reference genome(GenBank accession number: MH028738) and the assembled cp genome will be annotated using the online software GeSeq (Michael et al. Citation2017; Jin et al. Citation2020). Finally, the precisely annotated complete cp genome was submitted to GenBank with accession number MZ241297.
The length of the complete cp genome sequence of L. similis was 155,207 bp. It shows a single-circular molecule with a four-segment structure, consisting of a large single-copy (LSC, 88,994 bp) region, a small single-copy (SSC, 18,633 bp) region, and two inverted repeat (IRA and IRB) regions of 23,790 bp each. A total of 121 genes were predicted, including 77 protein-coding genes (PCGs), eight ribosomal RNA (rRNA) genes, and 36 transfer RNA (tRNA) genes. Among these assembled genes, all rRNAs, four PCGs (ycf2, rps7, ndhB, rps12) and seven tRNAs (trnN-GUU, trnR-ACG, trnA-UGC, trnI-GAU, trnV-GAC, trnL-CAA, and trnI-CAU) were with double copies. Intron-exon analysis showed the majority (103 genes, 85%) genes with no introns, whereas 18 genes (15%) contain introns.
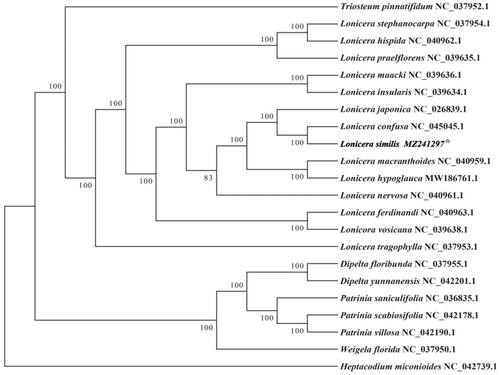
To make further research on the cp genome of L. similis, we choose mega7.0 software for alignment (https://www.megasoftware.net/). There were 21 cp genome sequences of Caprifoliaceae family (13 Lonicera species, three species from Patrinia genus, two species from Dipelta genus and the remaining of three species are from Heptacodium, Triosteum and Weigela) downloaded from GenBank to construct the phylogenetic tree through maximum-likelihood (ML) analysis. The ML tree based on GTR + gamma + I model was performed using RAxML (Version 8.0.19) with 1000 bootstrap replicates (Stamatakis Citation2014). We analyzed from the phylogenetic tree that L. similis belongs to genus Lonicera () and formed a nearly clade from L. confusa. It shows their relationship is the closest, and the support rate is 100%. Although L. japonica. is separate from L. similis, they still get a strong sister relationship. It also indicates we should make more comparative research between L. similis and other two species (L. confusa and L. japonica) to discover the potential economic value of L. similis.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The annotated chloroplast genome data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession number MZ241297. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA758746, SRR15665302, and SAMN21033719 respectively.
Additional information
Funding
References
- Cai ZC, Liu XH, Wang CC, Tan MX, Chen JL, Mei YQ, Wei LF, Chen H, Yang R, Chen JJ. 2020. [Research progress in molecular biology of Lonicerae Japonicae Flos]. Zhongguo Zhong Yao Za Zhi. 45(6):1272–1278.
- Dong W, Liu H, Xu C, Zuo Y, Chen Z, Zhou S. 2014. A chloroplast genomic strategy for designing taxon specific DNA mini-barcodes: a case study on ginsengs. BMC Genet. 15:138.
- Hu SQ, Dong GL, Chen XM, Huang LL, Yang X, Tong W, Bai LH. 2012. ITS sequence-based identification and utilization evaluation of “Nanjiang” (Lonicera similis Hemsl.), a local cultivar in Sichuan, China. Genet Resour Crop Evol. 59(4):547–555.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):1-11.
- Lv P, Fu YW, Chen XL, Zhang X. 2012. Study on Separation and Enriching of total flavonoids and chlorogenic acid from Lonicera Similes Hemsl. with Polyamide. J Chengdu Univ Tradit Chin Med. 35(04):52–54.
- Ma YY, Yang M, Zhou J, Li YC. 2007. Survey of Research on Lonicera japonica. Paper presented at: Proceedings of the 9th National Symposium on Traditional Chinese Medicine and Natural Medicine. Chinese Pharmaceutical Association; Academic Affairs Department of Chinese Pharmaceutical Association. p. 584–586.
- Michael T, Pascal L, Tommaso P, Elena SU, Axel F, Ralph B, Stephan G. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- National Pharmacopoeia Committee. 2015. The Pharmacopoeia of the People's Republic of China: One. The 2015 edition. Beijing: China Medical Science and Technology Press; 230–232.
- Saina JK, Li ZZ, Gichira AW, Liao YY. 2018. The complete chloroplast genome sequence of tree of heaven (Ailanthus altissima (Mill.) (Sapindales: Simaroubaceae), an important pantropical tree. IJMS. 19(4):929.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Weng YX, Chen XH, Liu ZH. 2011. Study on the chemical constituents of chlorogenic acids from the dried leaves of Lonicera similis Hemsl. Medicinal Plant. 2(10):56–58.
- Zeng AQ, Hua H, Chen CR, Liu L, Zhang M, Luo Y, Zhao JN. 2020. [Comparative study on anti-inflammatory effect of Lonicerae Japonicae Flos and Lonicerae Flos]. Zhongguo Zhong Yao Za Zhi. 45(16):3938–3944.
- Zhang X, Zou LH, He YL, Peng C, Guo L, Xiong L. 2018. Triterpenoid saponins from the buds of Lonicera similis. Nat Prod Res. 32(19):2282–2290.