Abstract
Zelkova sinica is a popular landscape plant in China because of its wide adaptation, strong disease resistance, large crown and beautiful fall color. Here, we assembled the complete chloroplast (cp) genome of Z. sinica based on genome skimming data. The cp genome is 158,924 bp in length including two copies of inverted region (IR, 26,427 bp) separated by the large single copy (LSC, 87,318 bp) and small single copy (SSC, 18,752 bp) regions. It encodes 111 unique genes, containing 77 protein-coding genes, 30 tRNA genes, and 4 rRNA genes, with 18 duplicated genes in the IR regions. Phylogenetic analysis shows that Z. sinica is sister to Z. schneideriana in Ulmaceae family.
Zelkova sinica Schneid., which belongs to Ulmaceae, is a species naturally distributed in the northwestern region in China (Chen and Huang Citation1999). Z. sinica is one of the most popular landscape plants in China due to its adaptability to varied environmental conditions, strong disease resistance, outstanding crown, and beautiful fall color (Jin and He Citation2005). Because of its beautiful and durable texture, the wood of this species has been widely used for making furniture (Jin and Hu Citation2012). Nowadays, Z. sinica is considered as a rare and endangered species because of a sharp decline of its distribution range, which is a consequence of overexploitation of the wild plants for commercial purposes and inefficient propagation methods (Fu and Jin Citation1992). However, researches regarding its genetic background are extremely scare. In this study, we used genome skimming data for assembling the complete cp genome of Z. sinica. The genome sequence was registered into GenBank with the accession number MW850753.
One individual of Z. sinica was sampled at Jinming Campus of Henan University (114°18′29.84″E, 34°49′13.53″N), Kaifeng, Henan, China. Fresh leaves were collected for genomic DNA extraction using modified CTAB (Cetyltrimethylammonium Bromide) reagents (Plant DNAzol, Shanghai, China) according to the manufacturer’s protocol. A voucher specimen (Luxian Liu LLX2021032506) was deposited at the Herbarium of Henan University. High quality DNA was sheared to yield fragments less than or equal to 800 bp, and the 500 bp short-insert length paired-end library was prepared and sequenced on an Illumina HiSeq X10 to obtain reads of 150 bp at Beijing Genomics Institute (BGI, Wuhan, China). The raw data was screened by quality with Phred score <30 and assembled into contigs using the CLC Genomic Workbench (CLC Inc. Aarhus, Denmark). The complete cp genome of Z. sinica was constructed and annotated using the software Geneious R11 (Biomatters, Auckland, New Zealand) following description in Liu et al. (Citation2017) and Liu et al. (Citation2020) with Z. serrata (GenBank accession number: MG717940) as a reference. Phylogenetic tree for 23 whole cp genome sequences of Rosales was constructed using maximum likelihood (ML) method implemented in RAxML-HPC v8.2.12 on the CIPRES cluster (Miller et al. Citation2010) with Cucumis sativus and Cucumis melo as outgroups. 1000 bootstrap iterations were conducted with other parameters using the default settings and GTR + G was determined by the software jModel Test v2.1.4 (Posada Citation2008) as the best-fit nucleotide substitution model.
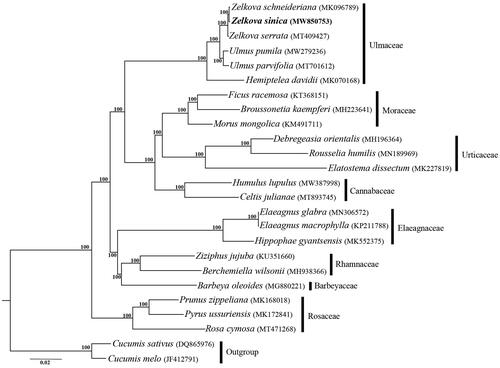
There were 118,113,846 paired-end reads received for Z. sinica, and 56,519,152 reads were removed from the raw data after trimming low quality sequences. The cp genome of Z. sinica was 158,924 bp in length, and had a typical quadripartite structure consisting of an 87,318 bp large single copy region (LSC), a 18,752 bp small copy region (SSC) and two 26,427 bp inverted repeats (IRs). Within the genome of Z. sinica, there are 111 unique genes, including 77 protein-coding genes, 30 tRNA genes, and 4 rRNA genes, additionally with 18 duplicated genes in the IR regions. Among the 111 genes, 6 tRNA genes, and 9 protein-coding genes contain a single intron, and three genes (rps12, clpP, and ycf3) contain two introns. The overall GC content of the total length, LSC, SSC, and IR regions is 35.6, 33.0, 28.3 and 42.3%, respectively. The phylogeny revealed that the three representative genera of Ulmaceae, including Zelkova, Ulmus and Hemiptelea, formed a full supported clade, and Z. sinica is sister to Z. schneideriana within Zelkova (). The result revealed in this study is consistent with that of Chase et al. (Citation2016) except for the position of Barbeyaceae, which is sister to the families including Dirachmaceae, Rhamnaceae, and Elaeagnaceae in the latter one.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MW850753. The associated Bio-Project, SRA and Bio-Sample numbers of the raw sequence data for assembling the cp genome are PRJNA735441, SRR14741978, and SAMN19582534, respectively.
Additional information
Funding
References
- Chase MW, Christenhusz MM, Fay MF, Byng JW, Judd WS, Soltis DE, Mabberley DJ, Sennikov AN, Soltis PS, Stevens PF. 2016. An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 181:1–20.
- Chen HY, Huang CG, 1999. Ulmaceae. In: Flora Republicae Popularis Sinicae. Vol. 22. Beijing, China: Science Press; p. 385.
- Fu LG, Jin JM. 1992. Plants red book of China-Rare and Endangered plants. Vol. I. Beijing, China: Science Press.
- Jin XL, He P. 2005. Biological characteristics in Zelkova. Nonwood for Res. 23:45–47.
- Jin XL, Hu XJ. 2012. Callus induction and plant regeneration from immature embryos of Zelkova sinica Schneid. Hort Sci. 47:790–792.
- Liu LX, Du YX, Folk RA, Wang SY, Soltis DE, Shang FD, Li P. 2020. Plastome evolution in Saxifragaceae and multiple plastid capture events involving Heuchera and Tiarella. Front Plant Sci. 11:361.
- Liu LX, Li R, James RW, Li X, Li P, Cameron KM, Fu CX. 2017. The complete chloroplast genome of Chinese bayberry (Morella rubra, Myricaceae): implications for understanding the evolution of Fagales. Front Plant Sci. 8:968.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gateway Computing Environments Workshop (GCE), 2010, IEEE; p.1–8.
- Posada D. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol. 25(7):1253–1256.