Abstract
We report here for the first time the complete plastid genome of Cheniella didyma of the legume family. The plastid genome has a typical circular structure with a total length of 157,186 bp and contains two inverted repeat regions (IRs, 24,455 bp), a large single-copy region (LSC, 89,410 bp), and a small single-copy region (SSC, 18,866 bp). This is the first report of the complete plastid genome sequence of Cheniella, a genus recently segregated from Bauhinia s.l. The phylogenetic analysis based on 77 coding regions of the plastome of this species and those of the related species strongly suggested that C. didyma is sister to Lysiphyllum and is not directly related to Bauhinia s.s.
Cheniella didyma (H.Y. Chen) R.Clark & Mackinder 2017, a woody liana with pure white flowers blossom from July to October, is a beautiful species of the genus Cheniella in the legume family (Clark et al. Citation2017). It occurs exclusively in Guangdong Province and Guangxi Zhuang Autonomous Region of China. We herein assembled and annotated for the first time the complete plastome of C. didyma using a method of genomic sequencing to provide genetic and genomic information for further systematic and genetic researches.
Leaf tissues of Cheniella didyma were taken from Gaoshuikeng village, Enping, Guangdong Province, China (112.07E, 22.17N). The specimens (vouchers: TuTY4691_9 contact: [email protected]) were deposited in the herbarium of South China Botanical Garden (IBSC), Guangzhou, China. We extracted the total genomic DNA by a modified CTAB method (Doyle and Doyle Citation1987). The isolated total genomic DNA was fragmented to make a library of 300-500 bp, and the paired-end sequences in length of ca. 150 bp were generated with Illumina (HiSeq X-Ten) at Beijing Genomics Institute (BGI) in Wuhan, China. The plastome was assembled by GetOrganelle pipeline (Bankevich et al. Citation2012; Langmead and Salzberg Citation2012; Wick et al. Citation2015; Jin et al. Citation2020), and Plastid Genome Annotator (PGA) (Qu et al. Citation2019) and Geneious (Kearse et al. Citation2012) were used to annotate and align the complete plastome. The annotated plastome has been deposited in GenBank (accession number: MZ230991).
To reconstruct the phylogenetic position of the species, we downloaded 13 plastid genome data of related species within Cercidoideae of the legume family from GenBank and used Cercis as outgroup to reconstruct the phylogenetic position of Cheniella didyma () (Sabir et al. Citation2014; Wang et al. Citation2017, Citation2018; Gu et al. Citation2019, Citation2020). We aligned the data matrix using MAFFT (Katoh and Standley Citation2013) as built in Geneious with default parameters. The phylogenetic relationship was estimated using the maximum likelihood method by RaxML-HPC2CIPRES Science Gateway (Miller et al. Citation2010) with models recommended by ModelFinder (Kalyaanamoorthy et al. Citation2017) based on a data matrix of concatenation of 77 coding regions (CDS). The branch supports were estimated using 1000 replicates of bootstrap. The complete plastid genome of C. didyma was 157,186 bp in length with a typical quadripartite structure: a large single copy (LSC) region of 89,410 bp and a small single copy (SSC) region of 18,866 bp, respectively. These two regions were separated by two inverted repeat regions (IRa and IRb), each of 24,455 bp in length. We recovered a total of 121 functional genes, including 80 protein-coding genes, 37 tRNA genes, and 4 rRNA genes. The overall GC content was 36.2%.
Figure 1. The maximum-likelihood (ML) phylogenetic tree based on 77 CDS of the plastid genomes. Numbers near the branches are bootstrap support values.
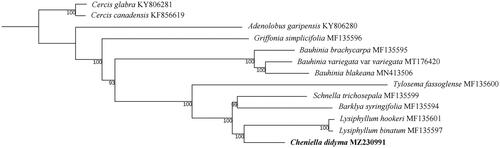
The phylogenetic analysis suggested that all the branches of the tree are strongly supported, suggesting the power of the plastid genome data in resolving the phylogenetic relationships within Cercidoideae. Cheniella didyma is recovered as a sister of Lysiphyllum and is not directly related to Bauhinia s.s, thus not conflicts with the treatment of Cheniella as a segregated genus by Clark et al. (Citation2017). It may be expected that a comprehensive sampling covering more species of Cheniella and related taxa (especially Phanera) in future shall shed more light on the phylogenetic relationships of this plant group.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The complete plastid genome of Cheniella didyma of this study is available in NCBI GenBank database (https://www.ncbi.nlm.nih.gov) with the accession number: MZ230991. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA746089, SRR15130292, and SAMN20180496.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Clark RP, Mackinder BA, Banks H. 2017. Cheniella gen. nov. (leguminosae: Cercidoideae) from southern China, Indochina and Malaysia. Eur J Taxon. 360:1–17.
- Doyle JJ, Doyle JL. 1987. A rapid isolation procedure for small amounts of leaf tissue. Phytochem Bull. 19:11–15.
- Gu SR, Chen Y, Zheng DJ, Meng SY, Tu TY. 2020. The complete plastid genome of Bauhinia variegata L. var. variegata (Leguminosae). Mitochondrial DNA Part B. 5(2):1701–1702.
- Gu SR, Lai Q, Zeng QB, Tu TY, Zhang DX. 2019. The complete plastid genome of Hong Kong Orchid Tree, Bauhinia × blakeana Dunn (Leguminosae). Mitochondrial DNA Part B. 4(2):3454–3455.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, Von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–357.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop, New Orleans, LA, November 2020; p. 1–8.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15:50.
- Sabir J, Schwarz EN, Ellison N, Zhang J, Baeshen NA, Mutwakil M, Jansen RK, Ruhlman TA. 2014. Evolutionary and biotechnology implications of plastid genome variation in the inverted-repeat-lacking clade of legumes. Plant Biotechnol J. 12(6):743–754.
- Wang YH, Wang H, Yi TS, Wang YH. 2017. The complete chloroplast genomes of Adenolobus garipensis and Cercis glabra (Cercidoideae, Fabaceae). Conservation Genet Resour. 9(4):635–638.
- Wang YH, Wicke S, Wang H, Jin JJ, Chen SY, Zhang SD, Li DZ, Yi TS. 2018. Plastid genome evolution in the early-diverging legume subfamily Cercidoideae (Fabaceae). Front Plant Sci. 9:138.
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31(20):3350–3352.
