Abstract
The 15,619 bp mitochondrial genome of Jebusaea hammerschmidtii was assembled from short reads, annotated, and compared to the genomes of other longhorn beetles (Cerambycidae). Gene content was typical of animal mitochondrial genomes and contained 13 protein-coding, 22 tRNA, and 2 rRNA genes. Gene organization was identical to that of other longhorn beetles. Phylogenetic analysis placed J. hammerschmidtii within the subfamily Cerambycinae, and strongly supported the monophyly of the Cerambycinae, Lamiinae, and Prioninae subfamilies.
Jebusaea hammerschmidtii (Reiche, 1878) (Coleoptera: Cerambycidae) is an important pest of the date palm (Phoenix dactylifera) in the Middle East and North Africa (El-Shafie et al. Citation2017). Females lay eggs at the base of palm fronds or in cracks on the tree trunk. After hatching, larvae bore into the tree, causing physical damage as they develop (El-Shafie Citation2015). As of May 2021, a single nucleotide record was available in GenBank for J. hammerschmidtii (COI, MG564344.1). Here we sequenced, assembled, and annotated the complete J. hammerschmidtii mitogenome and compared it to the mtDNAs of other longhorn beetles.
A single unsexed larva was obtained from a naturally infested date palm in the orchard of the Date Palm Research Center of Excellence at King Faisal University, Al-Ahsa, Saudi Arabia (25°16′04.8″N 49°42′25.2″E). The larva was identified as J. hammerschmidtii by specialists on the basis of its morphology and the fact that no other species with similar morphology occurs in this region. The entire specimen was utilized for DNA extraction and thus not deposited in a collection. DNA purification followed the ‘salting-out’ protocol (https://support.10xgenomics.com/permalink/7HBJeZucc80CwkMAmA4oQ2). DNA was cleaned using AMPure XP beads and 0.6 ng was utilized for barcoding and library construction using the Chromium Genome Reagent Kit Protocol v2 (RevB). The library was sequenced on a Illumina NextSeq 500 mid-output flow cell with 150 bp paired-end reads. Resulting fastq files were processed with LongRanger v2.2.2 (basic pipeline) (Zheng et al. Citation2016) to remove barcodes, then de-interleaved using ‘reformat.sh’ from BBMap v38.83 (Bushnell Citation2014). The mitogenome of another Cerambycinae beetle, Xylotrechus grayii (KM112084), was downloaded using ncbi-acc-download v0.2.5 (https://github.com/kblin/ncbi-acc-download) and used as mapping seed to identify mitochondrial sequences in the J. hammerschmidtii short-read dataset. We used two passes of GetOrganelle v1.7.3.2 (Jin et al. Citation2020a) to assemble the J. hammerschmidtii mitogenome (1st pass, ‘-w 111 -R 10 -F animal_mt’; 2nd pass, ‘-w 100 -R 15 -F animal_mt’). The first pass was used to identify all read pairs that matched the X. grayii mtDNA or the animal mtDNA database used by GetOrganelle. In the second round, mtDNA-matching read pairs were used as input and the entire mtDNA was recovered as a circular molecule of 15,619 bp. J. hammerschmidtii mtDNA was annotated with GeSeq v2.03 (Tillich et al. Citation2017) with available Cerambycinae mtDNAs as annotation references (Aeolesthes oenochrous, Massicus raddei, Neoplocaederus obesus, Epipedocera atra, Nortia carinicollis, X. grayii, Xystrocera globosa). ARWEN v1.2.3 (Laslett and Canbäck Citation2008) and tRNAscan-SE v.2.0.7 (Chan and Lowe Citation2019) were used for tRNA prediction with default settings. Redundant tRNA predictions were removed and the protein coding gene annotation was curated manually. Ribosomal RNA genes were extended after annotation with MITOS web server (commit 6b33f95) (Bernt et al. Citation2013). The final annotation revealed a gene content and organization identical to other Cerambycidae beetles, with 13 protein coding genes (PCGs), 2 ribosomal RNAs, and 22 transfer RNAs (Accession MZ054170).
Out of 12 PCGs for which start codons were identified, five used ATT (ATP8, ND2, ND3, ND4L, and ND5), three used ATG (COX3, CYTB, and ND4), two used ATC (COX2 and ND6), one used ATA (ATP6), and one used TTG (ND1). The start codon for COX1 could not be determined. The TAA stop codon was identified for 4 PCGs (ND4L, ND6, ATP6, and ATP8), and an additional 6 PCGs had an incomplete stop codon (T) hypothesized to form TAA by polyadenylation (ND2, ND4, ND5, COX1, COX2, and COX3). Three PCGs had a TAG stop codon (ND1, ND3, and CYTB). All tRNAs were predicted to form cloverleaf secondary structures.
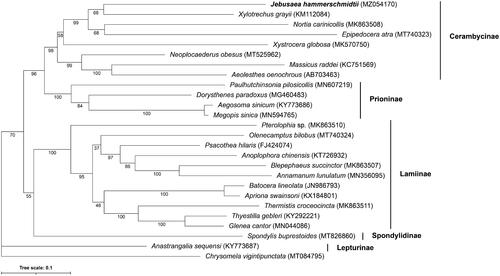
A total of 24 Cerambycidae mtDNA genomes plus an outgroup from the Chrysomelidae subfamily (Chrysomela vigintipunctata) were retrieved from RefSeq and used to place our J. hammerschmidtii mtDNA genome in a phylogenetic context (Kim et al. Citation2009; Chiu et al. Citation2016; Li et al. Citation2016; Wang et al. Citation2016; Jin et al. Citation2017; Liu et al. Citation2018; Que et al. Citation2019; Wang et al. Citation2019a, Citation2019b, Citation2019c, Dai et al. Citation2020; Li and Lu Citation2020; Lin et al. Citation2020; Su and Wang Citation2020; Yan et al. Citation2020; Jin et al. Citation2020b; Dong et al. Citation2021a, Citation2021b). Protein sequences from each mitogenome were concatenated and aligned using MAFFT v.7.455 (Katoh and Standley Citation2013), and a maximum likelihood phylogeny constructed using IQ-TREE v.2.1.2 (Minh et al. Citation2020) with the mtZOA+F + R5 model using C. vigintipunctata as the outgroup for rooting the tree. Node support was calculated after 10,000 ultrafast boostrap replicates. J. hammerschmidtii clusters with X. grayii with a bootstrap support of 68 (). Despite the low support for the grouping of J. hammerschmidtii and X. grayii, the clade containing J. hammerschmidtii, X. grayii, N. carinicollis, and E. atra has bootstrap support of 99. Monophyly of subfamily Cerambycinae, which contains J. hammerschmidtii, is recovered with bootstrap support of 98. The phylogeny supports monophyly of Prioninae and Lamiinae subfamilies with bootstraps of 100 in both cases, corroborating previous studies (Nie et al. Citation2021) (). Future analyses with denser taxon sampling will help elucidate the tribe-level phylogeny of longhorn beetles.
Figure 1. Maximum Likelihood phylogenetic tree of Cerambycidae beetle mtDNA genomes. Accession numbers are given after each species name. Numbers below each node represent ultrafast bootstrap support values after 10,000 replicates. The tree was rooted by setting Chrysomela vigintipunctata as the outgroup taxa.
Acknowledgements
We thank Julia Portocarrero and Magdy Alabady at the University of Georgia Genomics and Bioinformatics Core for assistance with library preparation and sequencing.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The annotated mitochondrial genome described here is available in GenBank under accession MZ054170 (https://www.ncbi.nlm.nih.gov/nuccore/MZ054170). Raw mitochondrial reads used for assembly are available in the SRA under accession SRR14321410 (https://www.ncbi.nlm.nih.gov/sra/SRR14321410).
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Bushnell B. 2014. BBMap: a fast, accurate, splice-aware aligner. Technical Report LBNL-7065E, Lawrence Berkeley National Lab. (LBNL), Berkeley, CA (United States).
- Chan PP, Lowe TM. 2019. tRNAscan-SE: searching for tRNA Genes in genomic sequences. In: Kollmar, M., editor, Gene prediction: methods and protocols, methods in molecular biology. New York (NY): Springer; p. 1–14.
- Chiu WC-H, Yeh W-B, Chen M-E, Yang M-M. 2016. Complete mitochondrial genome of Aeolesthes oenochrous (Fairmaire) (Coleoptera: Cerambycidae): an endangered and colorful longhorn beetle. Mitochondrial DNA Part A. 27(1):686–687.
- Dai X-Y, Zhang H, Xu X-D, Jia Y-Y, Zhang J-Y, Yu D-N, Cheng H-Y. 2020. The complete mitochondrial genome of Annamanum lunulatum (Coleoptera: Lamiinae) and its phylogeny. Mitochondrial DNA B Resour. 5(1):551–553.
- Dong Z, Wang X, Li Y, Huang D, Lu W. 2021a. Complete mitochondrial genome of an Asian longicorn beetle, Olenecamptus bilobus Fabricius (Coleoptera: Cerambycidae: Lamiinae). Mitochondrial DNA B Resour. 6(2):560–561.
- Dong Z, Yi X, Li S, Li Y, Hu D, Zheng X, Lu W. 2021b. Characterization of the complete mitochondrial genome of Epipedocera atra Pic (Cerambycidae: Cerambycinae: Tillomorphini). Mitochondrial DNA B Resour. 6(1):38–39.
- El-Shafie H. 2015. Biology, ecology and management of the longhorn date palm stem borer Jebusaea hammerschmidti (Coleoptera: Cerambycidae). Outlook Pest Man. 26(1):20–23.
- El-Shafie H, Abdel-Banat B, Al-Hajhoj M. 2017. Arthropod pests of date palm and their management. CAB Reviews. 12:49.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020a. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Jin M, Zwick A, Ślipiński A, Keyzer R. d, Pang H. 2020b. Museomics reveals extensive cryptic diversity of Australian prionine longhorn beetles with implications for their classification and conservation. Syst Entomol. 45(4):745–770.
- Jin Y, LiJun C, YongXin G, DanFeng W, Min C. 2017. Determination of the complete mitochondrial genome of Thyestilla gebleri and comparative analysis of the mitochondrial genome in Cerambycidae. Chin J Appl Entomol. 54(5):755–766.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kim K-G, Hong MY, Kim MJ, Im HH, Kim MI, Bae CH, Seo SJ, Lee SH, Kim I. 2009. Complete mitochondrial genome sequence of the yellow-spotted long-horned beetle Psacothea hilaris (Coleoptera: Cerambycidae) and phylogenetic analysis among coleopteran insects. Mol Cells. 27(4):429–441.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24(2):172–175.
- Li S, Lu W. 2020. The complete mitochondrial genome of an Asian longicorn beetle, Neoplocaederus obesus (Gahan) (Coleoptera: Cerambycidae: Cerambycinae). Mitochondrial DNA B Resour. 5(3):2684–2685.
- Li W, Yang X, Qian L, An Y, Fang J. 2016. The complete mitochondrial genome of the citrus long-horned beetle, Anoplophora chinensis (Coleoptera: Cerambycidae). Mitochondrial DNA Part A. 27(6):4665–4667.
- Lin B, Sun Y, Ma J, Deng R, Sheng L, Guan X. 2020. The complete mitochondrial genome of Spondylis buprestoides (Coleoptera: Cerambycidae). Mitochondrial DNA Part B. 5(3):3773–3774.
- Liu Y-Q, Chen D-B, Liu H-H, Hu H-L, Bian H-X, Zhang R-S, Yang R-S, Jiang X-F, Shi S-L. 2018. The complete mitochondrial genome of the Longhorn Beetle Dorysthenes paradoxus (Coleoptera: Cerambycidae: Prionini) and the implication for the phylogenetic relationships of the cerambycidae species. J Insect Sci. 18(2):12.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the Genomic Era. Mol Biol Evol. 37(5):1530–1534.
- Nie R, Vogler AP, Yang X-K, Lin M. 2021. Higher-level phylogeny of longhorn beetles (Coleoptera: Chrysomeloidea) inferred from mitochondrial genomes. Syst Entomol. 46(1):56–70.
- Que S, Yu A, Liu P, Jin M, Xie G. 2019. The complete mitochondrial genome of Apriona swainsoni. Mitochondrial DNA Part B. 4(1):931–932.
- Su R, Wang X. 2020. The complete mitochondrial genome of the Megopis sinica white (Coleoptera: Cerambycidae: Prioninae). Mitochondrial DNA Part B. 5(1):236–237.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wang J, Dai X-Y, Xu X-D, Zhang Z-Y, Yu D-N, Storey KB, Zhang J-Y. 2019a. The complete mitochondrial genomes of five longicorn beetles (Coleoptera: Cerambycidae) and phylogenetic relationships within Cerambycidae. PeerJ. 7:e7633.
- Wang J, Lan D-Y, Dai X-Y, Yu D-N, Storey KB, Zhang J-Y. 2019b. The complete mitochondrial genome of Xystrocera globosa (Coleoptera: Cerambycidae) and its phylogeny. Mitochondrial DNA Part B. 4(1):1647–1649.
- Wang X, Zheng X, Lu W. 2019c. The complete mitochondrial genome of an Asian longicorn beetle Glenea cantor (Coleoptera: Cerambycidae: Lamiinae). Mitochondrial DNA B Resour. 4(2):2906–2907.
- Wang Y-T, Liu Y-X, Tong X-L, Ren Q-P, Jiang G-F. 2016. The complete mitochondrial genome of the longhorn beetle, Massicus raddei. Mitochondrial DNA Part A. 27(1):209–211.
- Yan J, Li W, Song P, Li Y, Feng S, Liu D. 2020. Characterization of the complete mitochondrial genome of Chrysomela vigintipunctata (Coleoptera: Chrysomelidae). Mitochondrial DNA Part B. 5(2):1475–1476.
- Zheng GXY, Lau BT, Schnall-Levin M, Jarosz M, Bell JM, Hindson CM, Kyriazopoulou-Panagiotopoulou S, Masquelier DA, Merrill L, Terry JM, et al. 2016. Haplotyping germline and cancer genomes with high-throughput linked-read sequencing. Nat Biotechnol. 34(3):303–311.
