Abstract
Parnassius glacialis is a butterfly species distributed in China, Korea, Japan. The complete P. glacialis mitochondrial genome was assembled using Illumina sequencing data. The mitogenome is 15,353 bp long and contains 13 protein-coding genes, 22 transfer RNA genes, and 2 ribosomal RNA genes. A phylogenetic analysis of P. glacialis and 14 related Papilionidae species indicated that P. glacialis is clustered with other Parnassius species. This study generated useful genetic information for future studies on the taxonomy, phylogeny, and evolution of Papilionidae species.
Parnassius is a genus of Papilionidae (Insecta: Lepidoptera). In contrast to most Parnassius species, which live in high altitude plateau regions, Parnassius glacialis is widely distributed at relatively low altitudes in China (Wang et al. Citation2019). The body of this butterfly species is black and covered with long yellow hair, and its wings are translucent with brown veins (Wu Citation1998a; Citation1998b; Hao et al. Citation2017). Molecular systematics, molecular evolution, population genetics, and phylogenetic studies of butterflies have commonly used COI, COII, 16S rDNA, and other molecular markers (Wang et al. Citation2013; He et al. Citation2016; Qin Citation2017; Wang Citation2017). Additionally, earlier investigations on P. glacialis involved ecological surveys as well as analyses of larvae and phylogeography (Ding et al. Citation2007; Akiyama and Nishida Citation2013; Wang Citation2017). Wang (Citation2017) elucidated the genetic diversity of P. glacialis by analyzing mitogenomic AT-rich sequences. However, the P. glacialis whole-mitogenome characteristics remain relatively uncharacterized. To date, in the genus Parnassius, the complete mitogenome has been sequenced for only nine species, including Parnassius apollonius, Parnassius mercurius, and Parnassius apollo (Dechaine and Martin Citation2004; Chen et al. Citation2014; Zhang et al. Citation2017). In this study, we sequenced and annotated the complete P. glacialis mitogenome to further clarify the mitochondrial characteristics and evolutionary history of Papilionidae species.
In this study, the voucher specimen of P. glacialis was collected from Chengkou, Chongqing, China (31.816 N, 109.038 E; specomen voucher number CQ1151m-2). The voucher specimen was preserved at −20 °C and deposited at Chongqing Normal University, China (https://www.cqnu.edu.cn/, Yaping Hu: [email protected]). The genomic DNA was extracted from three legs of a single individual using TIANamp Genomic DNA Kit (TIANGEN, Beijing, China). The sequencing library was produced using the Illumina Truseq™ DNA Sample Preparation Kit (Illumina, San Diego, USA) according to the manufacturer's recommendations. The prepared library was loaded on the Illumina Novaseq 6000 platform for PE 2 × 150 bp sequencing at Novogene (Beijing, China). The raw data were used to assemble the complete cp genome using the GetOrganelle pipeline (Jin et al. Citation2020). Genome annotation was performed with Mitoz annotation module (Meng et al. Citation2019). The annotated genome sequence was deposited in GenBank under Accession Number MZ353680.
The circular mitogenome of P. citrinariu was 15,353 bp in length, with 40.75% A, 40.36% T, 11.32% G, and 7.57% C, the higher value of A + T content (80.17%) compare to G + C content (19.83%), indicating there was high A + T bias in P. glacialis. 37 genes were predicted, including 13 protein-coding genes, 22 transfer RNAs, and two ribosomal RNAs genes.
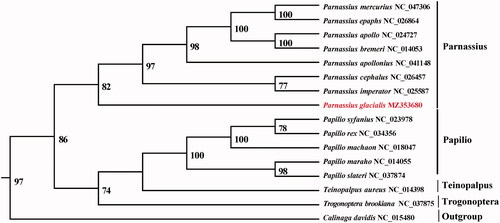
A phylogenetic analysis was performed using complete mitogenomes from 14 Papilionidae species with Calinaga davidis serving as the outgroup taxa. The genomes were aligned with the MAFFT v7.388 using default settings (Katoh and Standley Citation2013). The phylogenetic analysis was conducted based on maximum likelihood (ML) analyses implemented in IQ-TREE v2.1.2 with the GTR + F+R2 nucleotide substitution model, which was selected by ModelFinder (Kalyaanamoorthy et al. Citation2017; Minh et al. Citation2020). The support for the inferred ML tree was inferred by bootstrapping with 1000 replicates. The analysis shows that P. citrinariu in a clade with other Parnassius species (). This study provides important sequence information for species identification, and its phylogenetic relationship in Papilionidae species.
Figure 1. Maximum-likelihood (ML) tree based on 14 mitogenome sequences of representative butterflies that are in Papilioninae as ingroup and Calinaga davidis was designated as outgroup. Numbers on the nodes are bootstrap values based on 1000 replicates. The P. glacialis genome was marked in bold and red font.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov/) under the accession number MZ353680. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA762526, SAMN21397441 and SRR15858608, respectively.
Additional information
Funding
References
- Akiyama K, Nishida T. 2013. Highly-enhanced larval growth during the cold season mediated by the basking behavior of the butterfly Parnassius citrinarius (Lepidoptera: Papilionidae). Entomol Sci. 16(3):284–290.
- Chen YH, Huang DY, Wang YL, Zhu CD, Hao JS. 2014. The complete mitochondrial genome of the endangered Apollo butterfly, Parnassius apollo (Lepidoptera: Papilionidae) and its comparison to other Papilionidae species. J Asia-Pac Entomol. 17(4):663–671.
- Dechaine EG, Martin AP. 2004. Historic cycles of fragmentation and expansion in Parnassius smintheus (papilionidae) inferred using mitochondrial DNA. Evolution. 58(1):113–127.
- Ding L, Zhang YZ, Zhu CD. 2007. Preliminary study on taxonomy and phylogeny of Zerynthiini and Parnassiini (Lepidoptera, Papilionidae). Acta Zootaxonomica Sinica. 32(2):355–362.
- Hao XY, Mao ZH, Ren H, Tao RS. 2017. Analysis of geometric morphology of vein of Pamassius gutcmhs in different geographic populations. J Anhui Agric Sci. 45(34):84–88.
- He HY, Yu WD, Jiang WB. 2016. Research progress in mitochondrial genomics of butterflies. Chin Bull (Life Sci). 28 (9):978–985. .
- Jin JJ, Yu WB, Yang JB, Song Y, Depamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):1–31.
- Kalyaanamoorthy S, Minh BQ, Wong TK, Von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Meng GL, Li YY, Yang CT, Liu SL. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids RES. 47(11):63.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, Von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534.
- Qin JM. 2017. The study on mitochondrial genomes and molecular phylogenetics of butterflies [M]. Vol. 18. Beijing: Science Press.
- Wang W, Meng ZQ, Shi FX, Li FB. 2013. Advances in comparative mitogenomic studies of Lepidoptera (Arthropoda: Insecta). Chin Sci Bull. 58(30):3017–3029.
- Wang YL, Pan ZQ, Chen KK, Tao RS, Su CYg, Hao JS, Yang Q. 2019. Genetic differentiation and phylogeography of the alpine butterfly Parnassius glacialis (Papilionidae: Parnassinae) in China: evidence from mitogenomic AT-rich region. Acta Ent Sin. 62(4):475–488.
- Wang YL. 2017. Mitogenomic phylogeny of the main lineages of butterflies (Lepidoptera: Arthropoda: Insecta) and the Phylogeography of Parnassius glacialis based on the mitochondrial D-loop regions. PhD dissertation, Anhui Normal University, Wuhu, Anhui.
- Wu Q. 1998a. Parnassius glacialis (lower part). China Nat. 4:19–23.
- Wu Q. 1998b. Parnassius glacialis (upper part). China Nat. 3:17–20.
- Zhang M, Zhao P, Yin J, Li T, Zhang TT, Cao TW, Ma EB. 2017. Characterization of the complete mitochondrial genome of Parnassius nomion (Lepidoptera: Parnassiidae) and analysis of the higher-level phylogenetic relationships of butterflies based on mitochondrial genome. Acta Ent Sin. 60(11):1324–1338.
