Abstract
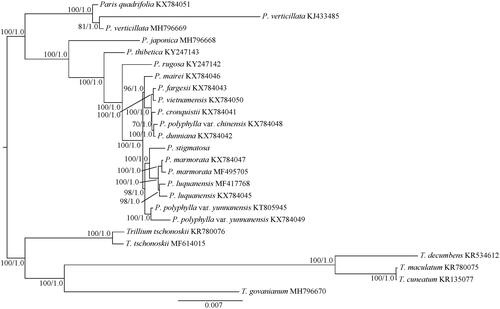
Paris stigmatosa is a new described species of Melanthiaceae. In this study, the complete chloroplast (cp) genome sequence of P. stigmatosa was first reported and characterized. The cp genome is 165,623 bp in length and contains a pair of inverted repeats (IRs, 34,165 bp) separated by a large (84,327 bp) and small (12,966 bp) single-copy regions. A total of 113 genes were predicted, including 79 protein-coding genes, 30 tRNA genes and 4 rRNA genes. The phylogenetic analysis suggested that P. stigmatosa is sister of the clade formed by P. marmorata and P. luquanensis.
The genus Paris (Melanthiaceae) comprises 27 species accepted in The Plant List (Citation2013) and divides into two subgenera Paris s. s. and Daiswa (Huang et al. Citation2016). Paris is well known in China for its medicinal qualities. In recent years, some new Paris species were gradually reported, such as P. lihengiana (Xu et al. Citation2019), P. tengchongensis (Ji et al. Citation2017) and P. nitida (Wang et al. Citation2017). Paris stigmatosa Shu D. Zhang is a new species described in 2008 (Zhang et al. Citation2008). This species is very similar to P. polyphylla in the morphology, but it has longer stigmas (21-34 mm). Paris stigmatosa is only geographically found in Yaoshan Mountain of Yunnan Province, China (Zhang et al. Citation2008). Until now, little information is known about this species. To better understand and utilize this species, we sequenced and analyzed the complete chloroplast (cp) genome of P. stigmatosa using high-throughput sequencing technology.
The specimen (03-1681) was collected from Yaoshan Mountain (Qiaojia, Yunnan, China; 103°00′E, 26.52′N) and deposited at Herbarium, Kunming Institute of Botany, CAS (KUN, http://www.kun.ac.cn/, Tao Deng, [email protected]). Genomic DNA was extracted with a modified CTAB (Cetyl Trimethyl Ammonium Bromide) method (Yang et al. Citation2014) from the fresh leaves. Purified DNA was fragmented and used to construct short-insert (350 bp) library using NEB Next Ultra DNA Library Prep Kit for Illumina (NEB, USA) as per manufacturer’s recommendations. Approximately 6 Gb raw data of 150 bp paired-end reads were generated using the Illumina HiSeq X ten platform at Beijing Novogene Bioinformatics Technology Co., Ltd. (Nanjing, China) and used for the cp genome assembly using SPAdes (Bankevich et al. Citation2012). The cp genome annotation was accomplished using PGA (Qu et al. Citation2019) with the cp genomes of P. marmorata (KX784047) and P. thibetica (KY247143) as reference sequences coupled with manual check and adjustment. The circle cp map of P. stigmatosa was generated by OGDRAW (Greiner et al. Citation2019).
The complete cp genome of P. stigmatosa (accession number MN723866) is 165,623 bp in length with a typical quadripartite structure containing two inverted repeats (IRs) of 34,165 bp, a large single copy (LSC) region of 84,327 bp and a small single copy (SSC) region of 12,966 bp. The overall GC content of the cp genome is 36.8%. A total of 113 unique genes consist of 79 protein-coding genes, 30 transfer RNA (tRNA) genes, and 4 ribosomal RNA (rRNA) genes, which is little different from other species of Paris (Huang et al. Citation2016). Among these genes, 15 genes (atpF, ndhA, ndhB, petB, petD, rpl16, rpl2, rpoC1, rps16, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, trnV-UAC) contain one intron and three genes (clpP, rps12 and ycf3) have two introns.
In this study, we constructed the phylogenetic tree and analyzed the phylogenetic position of P. stigmatosa based on the maximum likelihood (ML) and Bayesian inference (BI) methods (Ronquist et al. Citation2012; Stamatakis Citation2014). Five species from Trillium (T. tschonoskii, T. decumbens, T. maculatum, T. cuneatum and T. govanianum) were used as the outgroups. The cp genomes of P. stigmatosa and previously published species of Paris were used for phylogenetic analysis. The complete cp genome sequences were aligned by using MAFFT version 7.308 (Katoh and Standley Citation2013). The best-fit model (TIMef) for the dataset was determined by MODELTEST v.3.7 (Posada and Crandall Citation1998) with the Akaike Information Criterion (AIC) (Posada and Buckley Citation2004). BI was performed with Mrbayes v.3.2 (Ronquist et al. Citation2012). Two independent Markov Chain Monte Carlo (MCMC) chains were run, each with three heated and one cold chain. Each chain started with a random tree, default priors and sampling trees every 100 generations, with the first 25% discarded as burn-in. Stationarity was considered to be reached when the average standard deviation of split frequencies was <0.01. The ML analysis was performed with RAxML v.8.2.4 (Stamatakis Citation2014). The ML tree was inferred with the combined rapid bootstrap (1,000 replicates) and search for ML tree (the ‘-f a’ option). The GTRGAMMA model was used in the analysis as suggested (RAxML manual). The phylogenetic analysis showed that Paris is monophyletic and can be divided into two separate clades (), corresponding to the subgenera Paris s. s. and Daiswa (Huang et al. Citation2016). In Daiswa clade, P. marmorata and P. luquanensis are united in one clade and P. stigmatosa is positioned as a sister group to this clade ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov (https://www.ncbi.nlm.nih.gov/) under the accession no. MN723866. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA724898, SRR14420049, and SAMN19009008 respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64.
- Huang Y, Li X, Yang Z, Yang C, Yang J, Ji Y. 2016. Analysis of complete chloroplast genome sequences improves phylogenetic resolution in Paris (Melanthiaceae). Front Plant Sci. 7:1–12.
- Ji Y, Yang C, Huang Y. 2017. A new species of Paris sect. Axiparis (Melanthiaceae) from Yunnan, China. Phytotaxa. 306(3):234–236.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Posada D, Buckley TR. 2004. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst Biol. 53(5):793–808.
- Posada D, Crandall KA. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics. 14(9):817–818.
- Qu X, Moore M, Li D, Yi T. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):1–12.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, H€Ohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- The Plant List. 2013. Version 1.1. Published on the Internet; http://www.theplantlist.org/
- Wang Z, Cai X-Z, Zhong Z-X, Xu Z, Wei N, Xie J-F, Hu G-W, Wang Q-F. 2017. Paris nitida (Melanthiaceae), a new species from Hubei and Hunan, China. Phytotaxa. 314(1):145–149.
- Xu Z, Wei N, Tan Y, Peng S, Ngumbau VM, Hu G, Wang Q. 2019. Paris lihengiana (Melanthiaceae: Parideae), a new species from Yunnan, China. Phytotaxa. 392(1):45–53.
- Yang J, Li D, Li H. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Resour. 14(5):1024–1031.
- Zhang S, Wang H, Li D. 2008. A new species of Paris (Melanthiaceae) from Northeastern Yunnan, China. Novon. 18(4):550–554.