Abstract
We report the complete mitochondrial genomes of two antipatharian species, Stichopathes sp. SCBUCN-8849 and Stichopathes sp. SCBUCN-8850, collected between 120 and 180 m depth off Rapa Nui (∼ −27.1°, −109.4°). The size of the two mitogenomes are 20,389 bp (29.0% A, 15.2% C, 19.9% G, and 35.9% T) and 20,463 bp (29.0% A, 15.3% C, 19.9% G, and 35.8% T), respectively. Both mitogenomes have the classic Hexacorallia gene content of 13 protein-coding, two rRNA, and two tRNA genes plus a COX1 intron with embedded HEG as found in the Antipathidae and other antipatharian families.
The order Antipatharia includes over 279 described species in seven families that occur in all oceans between 2 and 8600 m (Molodtsova and Opresko Citation2017). Antipathidae is composed of eight genera distinguished by morphological differences in colony branching pattern, polyps, and skeletal spines (Molodtsova and Opresko Citation2021). Recent phylogenetic studies based on the internal transcriber spacer 1 and 2 (ITS1 and ITS2) rDNA (Bo et al. Citation2012) and mitochondrial genomes (Barrett et al. Citation2020) indicate that Antipathidae and several genera within Antipathidae are not monophyletic, highlighting the need for future taxonomic revisions.
During benthic surveys off Rapa Nui (Easter Island) two specimens of whip antipatharians with distinct colorations and morphologies (Supp. Mat.) were collected by chance, entangled in the propellers of a remotely operated vehicle. Specimen collection was performed under permissions Res. Ext N°41/2016 and N°3314/2017 from SUBPESCA (National Fishing Authority of Chile) to Universidad Católica del Norte (UCN) and from the “Consejo del Mar de Rapa Nui” (Rapa Nui Council of the Sea). A yellow morphotype was collected between 120 and 180 m (27.100° S; 109.431° W) and a red morphotype was collected at 180 m (27.101° S; 109.426° W). Specimens were stored in 95% ethanol and deposited in the Sala de Colecciones, UCN (SCBUCN, Javier Sellanes, [email protected]): SCBUCN-8849 (yellow) and SCBUCN-8850 (red). On both specimens, polyp arrangement was in a single row on only one side of the corallum, a diagnostic morphological character for the genus Stichopathes (Bo and Opresko Citation2015). Although morphological differences in skeletal spines characters, which are considered diagnostic for antipatharian species, were recorded (Supp. Mat.), neither could be assigned to described species by the authors or taxonomic experts consulted because of the poor quality or missing type material for many Stichopathes species, most of which were described over a century ago with limited information (Bo et al. Citation2012).
Isolated genomic DNA was submitted to Biopolymers Facility at Harvard Medical School for library preparation and next-generation sequencing (NextSeq 500). Trimmed reads (Trimmomatic-0.32, Bolger et al. Citation2014) were assembled de novo with SPAdes (Bankevich et al. Citation2012) on the University of New Hampshire Bioinformatics Core facility ron server. After circularizing and editing overlapping ends of the SPAdes contig in Geneious Prime 2021.1.1 (https://www.geneioius.com), trimmed reads (BBDuk v. 38.84) were mapped to the resulting reference sequence to generate a consensus sequence. Genes were annotated by manually adjusting Stichopathes luetkeni (NC018377) annotations mapped to the consensus sequence. Maximum-likelihood phylogenetic reconstructions were based on the concatenated mitochondrial protein-coding genes of these two Stichopathes specimens, 21 representatives of Antipatharia, and 8 representatives (outgroups) from Hexacorallia aligned with default MUSCLE (Edgar Citation2004) parameters. See Supp. Mat. for extended methods, including of ITS1-based reconstruction.
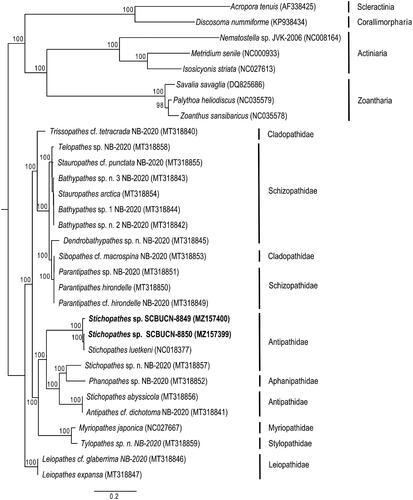
Both mitogenomes presented the classic pattern expected for Hexacorallia of 13 protein-coding genes, two rRNA genes, and two tRNA genes (tRNA-met and tRNA-trp) plus a COX1 intron with embedded HEG as found in Antipathidae and other antipatharian families (Barrett et al. Citation2020). The complete mitogenome of Stichopathes sp. SCBUCN-8849 (MZ157400) has 20,389 bp (29.0% A, 15.2% C, 19.9% G, and 35.9% T), whereas Stichopathes sp. SCBUCN-8850 (MZ157399) has 20,463 bp (29.0% A, 15.3% C, 19.9% G, and 35.8% T). These species were 98.71% similar (uncorrected p distances) across the entire mitogenome and 98.69% similar across the protein-coding genes. They form a well-defined clade with S. luetkeni and are sister to a clade containing the other three Antipathidae species and an Aphanipathidae species (). Across the mitochondrial proteome, Stichopathes sp. SCBUCN-8849 and SCBUCN-8850 were respectively 99.25% and 98.25% similar to S. luetkeni. Based on ITS1 rDNA sequences, the Rapa Nui Stichopathes species were genetically distinct from each other (38.27% uncorrected p, MZ450123-MZ450124) based on 343-bp alignment of the ITS1 region of 70 antipatharians and a scleractinian outgroup (Supp. Mat.). In the ITS1-based reconstruction, Stichopathes sp. SCBUCN-8849 groups within a clade mainly composed of species of the genus Cirrhipathes that differ from SCBUCN-8849 by 0.9-2.2%, whereas Stichopathes sp. SCBUCN-8850 groups within a clade (clade C in Bo et al. Citation2012) mainly composed of Antipathes species that differ from SCBUCN-8850 by 1.0–2.0%. These findings highlight the need for further phylogenetic studies and taxonomic revisions within Antipathidae. With only a few complete antipatharian mitogenomes available, the distinction of clades even at the family level is hindered (Barrett et al. Citation2020). Yet, phylogenetic information based on mitochondrial genomes is critical to understand evolutionary history and phylogeography and to provide insight into which taxa to prioritize for taxonomic revisions. A better understanding of their identity, evolutionary history, and distribution would, in turn, provide essential information for their conservation and management, especially in marine protected areas around remote islands, such as Rapa Nui. In addition, the mitogenomes presented here have potential implications for future biomedical research because antipatharians have been used for medicinal purposes in many cultures (Wagner et al. Citation2012).
Figure 1. Maximum-likelihood, phylogenetic reconstruction based on the complete mitochondrial proteome of two Stichopathes species (in bold) collected from Rapa Nui (Easter Island) and 21 representatives of Antipatharia and eight representatives from other Hexacorallia subclasses as outgroups: Scleractinia, Corallimorpharia, Actiniaria, and Zoantharia. Species names and GenBank accession numbers are included in parentheses at the tips. All nodes had bootstraps ≥ 76, but values were excluded in large, densely branched clades. See Supplemental data for details.
Supplemental Material
Download MS Word (1.9 MB)Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Data availability statement
The mitochondrial genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession no. MZ157399-MZ157400. The associated voucher repository (Sala de Colecciones, UCN, Javier Sellanes, [email protected]) numbers are SCBUCN-8850 and SCBUCN-8849 respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Barrett NJ, Hogan RI, Allcock AL, Molodtsova T, Hopkins K, Wheeler AJ, Yesson C. 2020. Phylogenetics and mitogenome organisation in black corals (Anthozoa: Hexacorallia: Antipatharia): an order-wide survey inferred from complete mitochondrial genomes. Front Mar Sci. 7:440.
- Bo M, Opresko DM. 2015. Redescription of Stichopathes pourtalesi Brook, 1889 (Cnidaria: Anthozoa: Antipatharia: Antipathidae). Breviora. 540:1–18.
- Bo M, Bavestrello G, Barucca M, Makapedua DM, Poliseno A, Forconi M, Olmo E, Canapa A. 2012. Morphological and molecular characterization of the problematic whip black coral genus Stichopathes (Hexacorallia: Antipatharia) from Indonesia (North Sulawesi, Celebes Sea). Zool J Linn Soc. 166:1–13.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797.
- Molodtsova T, Opresko D. 2017. Black corals (Anthozoa: Antipatharia) of the Clarion-Clipperton fracture zone. Mar Biodiv. 47(2):349–365.
- Molodtsova T, Opresko D. 2021. World List of Antipatharia. Antipathidae Ehrenberg, 1834. [accessed 2021 Mar 9]. Available at http://www.marinespecies.org/aphia.php?p=taxdetails&id=103301.
- Wagner D, Luck DJ, Toonen RJ. 2012. The biology and ecology of black corals (Cnidaria: Anthozoa: Hexacorallia: Antipatharia). Adv Mar Biol. 63:67–132.
