Abstract
Tetradium daniellii (Benn.) T. G. Hartley is an important medicinal, ornamental, and timber tree species and belongs to genus Tetradium in family of Rutaceae. It is widely distributed in warm temperate deciduous broad-leaved forest areas in northern China, Korean Peninsula and Japan. In this study, we sequenced its sample and determined complete chloroplast genome. The CP genome of T. daniellii has a circle structure with the length 158,446 bp, includes a small single copy region (17, 972 bp), a large single copy (86, 478 bp) and two inverted repeats (26,998 bp). There were 131 genes, which included 86 protein-coding genes, 8 rRNA and 37 tRNA, and overall GC content covered by 38.3%. The gene trnK-UUU, rps16, trnG-UCC, atpF, rpoC1, trnL-UAA, trnV-UAC, petB, petD, rpl16, rpl2, ndhB, trnI-GAU, trnA-UGC and ndhA contained an intron; gene clpP, ycf3 contained 2 introns. The phylogenetic result showed that T. daniellii had the closest relationship with Tetradium ruticarpum (NC_052830).
Tetradium daniellii (Benn.) T. G. Hartley is a plant of the genus Tetradium in the family Rutaceae, which is widely distributed in warm temperate deciduous broad-leaved forest areas in northern China, Korean Peninsula, and Japan, at present, the research of T. daniellii mainly focuses on Chemical Components (Yoo et al Citation2002; Lee et al Citation2011; Li et al Citation2013). However, we can hardly find the research about the complete chloroplast of T. daniellii, and in NCBI we can not find the record of the complete chloroplast genome sequence of it before this study. So in this study, we sequenced the sample and determined the complete chloroplast genome, then we did the phylogenetic analysis to determine the position in family Rutaceae.
The sample of T. daniellii was from Taishan Mountain, Taian, Shandong Province, China (N37°11′50″, E121°40′50″), we only collected several stems and leaves of the sample but not the whole plant. We used part of leaves to extract the DNA and stored others in National Plant Specimen Resource Center (http://www.cvh.ac.cn/, barcode SDF1004500, Lei Wang, Email: [email protected]). The fresh leaves were used to extract the total genomic DNA with the modified CTAB method (Doyle and Doyle Citation1987), then we constructed the library with an average length of 350 bp using the NexteraXT DNA Library Preparation Kit (Illumina, San Diego, CA) and sequenced by Illumina Novaseq 6000 platform. The 1.57 Gb clean sequence data was assembled by de novo assembler SPAdes v3.11.0 (Bankevich et al. Citation2012) with the reference genome Phellodendron amurense (NC_035551). Finally the complete chloroplast genome was annotated by PGA (Qu et al. Citation2019). We submitted the assembled genome DNA to GenBank under the accession number of MZ145060, and SRA submitted to NCBI under the BioProject No. PRJNA732282 and SRA number: SRR14663312
The chloroplast genome of T. daniellii has a circle structure with the size of 158,446 bp in length that contains a large single copy (LSC: 86,478 bp) region, a small single copy (SSC: 17,972 bp) and two inverted repeats (IRs: 26,998 bp) region. The overall GC content was 38.3%. There were 131 genes including 86 protein-coding genes, 37 tRNA and 8 rRNA. Each of trnK-UUU, rps16, trnG-UCC, atpF, rpoC1, trnL-UAA, trnV-UAC, petB, petD, rpl16, rpl2, ndhB, trnI-GAU, trnA-UGC and ndhA contained one intron respectively, clpP and ycf3 had two introns respectively. While rps12 had Trans splicing.
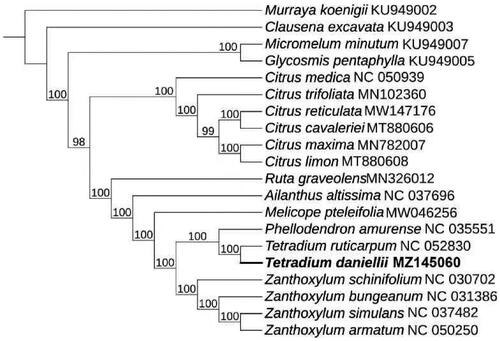
To determine the phylogenetic position of T. daniellii in family Rutaceae, we selected 19 complete chloroplast genomes from NCBI and aligned with T. daniellii by using Mafft 7.473 (Katoh and Standley Citation2013) with strategy of FFT-NS-2. Then we used model finder to selecte TVM + F + I + G4 model (Subha et al. Citation2017) and constructed the phylogenomic tree () by IQtree 2.0 (Minh et al. Citation2020) with 1000 bootstrap and Maximum-likehood method. During the ML tree construction, the complete chloroplast genomes of Murraya koenigii (MT806177) was used as outgroup. Then the result showed that T. daniellii had the closest relationship with Tetradium ruticarpum (NC_052830)
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MZ145060. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA732282, SRR14663312, and SAMN19316631 respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534..
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Lee DG, Choi K, Lee SH. 2011. GC/MS analysis of volatile constituents from woody plants. Appl Biol Chem. 38(4):723–730.
- Li W, Sun YN, Yan XT, Yang SY, Kim E-J, Kang HK, Kim YH. 2013. Coumarins and lignans from Zanthoxylum schinifolium and their anticancer activities. J Agric Food Chem. 61(45):10730–10740.
- Qu X-J, Moore MJ, Li D-Z, Yi T-S. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15:50.
- Subha K, Minh BQ, Wong TK, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589..
- Yoo SW, Kim JS, Kang SS, Son KH, Chang HW, Kim HP, Bae K, Lee CO. 2002. Constituents of the fruits and leaves of Evodia daniellii. Arch Pharmacal Res. 25(6):30–824.