Abstract
The mitochondrial genome of the spectacled parrotbill Sinosuthora conspicillata is sequenced by the Sanger method. The genome is 16,982 bp in length, comprising of 13 protein-coding genes (PCGs), 2 rRNA genes, 22 tRNA genes, 1 control region (D-loop), and 1 pseudo-control region. The PCGs of COX1 and ND3 use GTG and ATA as their starting codon, respectively, while all other PCGs start with ATG codons. Four PCGs (COX3, ND4, ND5, and ND6) are terminated with CCT, TAT, AGA, and TAG, respectively, and all other PCGs end with TAA. The 22 tRNAs range from 66 bp (tRNA-Ser) to 75 bp (tRNA-Leu) in length. The two rRNAs are 984 bp (12S) and 1600 bp (16S) in length. Phylogenetic analysis indicated that S. conspicillata is closely related to the congeneric vinous-throated parrotbill S. webbiana. This mitochondrial genome sequence offers a valuable resource for future conservation genetic and phylogenetic studies of birds in the family Sylviidae (Passeriformes).
The spectacled parrotbill (Sinosuthora conspicillata) is endemic to central and western China. It belongs to the family Sylviidae, with a synonym of Paradoxornis conspicillatus. It is uncommon in mountainous areas between 1360 and 2900 m, descending to 1000 m in winter (MacKinnon et al. Citation2000). While typically found alone, S. conspicillata sometimes associates with groups of vinous-throated parrotbills S. webbiana. In this study, we sequenced and analyzed the complete mitochondrial genome of S. conspicillata to provide insights into the phylogenetic relationships within the Sylviidae.
We set up a mist net at an elevation of 2645 m in the Sichuan Tangjiahe National Nature Reserve on 17 May 2015. After catching a spectacled parrotbill, we quickly bled its left cutaneous ulnar vein with a 10 uL blood capillary (blood sample code: CIBwj2015051702, deposited in the herpetological museum of Chengdu Institute of Biology by Jie Wang, [email protected]) and then released the bird at the capture site (E104.79877, N32.64463). Total DNA was extracted using the Tiangen DNA Extraction Kit (Beijing), amplified using 23 pairs of primers, and then sequenced using an ABI 3730 automatic sequencer. The complete sequence of mitochondrial DNA was assembled with DNAstar v7.1 (Burland Citation2000) and annotated using the MITOS Web Server (Bernt et al. Citation2013).
We used Circos software (Krzywinski et al. Citation2009) to show the structure of mitochondrial genome. It is 16,982 bp in length and includes 37 genes (two rRNA genes, 13 protein-coding genes/PCGs, and 22 tRNA genes). One PCG (ND6), two rRNA (12S and 16S), and 8 tRNAs (Gln, Ala, Asn, Cys, Tyr, Ser, Pro, and Glu) are on the light chain, whereas the other genes are on the heavy chain. The content of A + T is 54.18% (A, 29.73%, T, 24.45%, G, 14.56% and C, 31.26%). Two PCGs (COX1 and ND3) have GTG and ATA as the start codon, respectively, while all other PCGs start with ATG. Four PCGs (COX3, ND4, ND5, and ND6) use CCT, TAT, AGA, and TAG as the stop codon, respectively, while all others end with TAA. The 22 tRNAs range from 66 (tRNA-Ser) to 75 bp (tRNA-Leu) in size. Two rRNAs are 984 bp (12S) and 1600 bp (16S) long. The arrangement of genes is identical to S. webbiana, Suthora fulvifrons, Psittiparus gularis, and P. heudei (Zhang et al. Citation2015, Wen et al. Citation2017, He Citation2019, and Chen Citation2020).
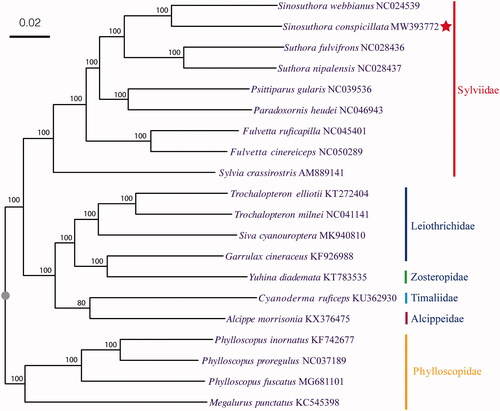
Using the great tit Parus major as an outgroup, we ran MrBayes 3.2.7a (Ronquist and Huelsenbeck Citation2003) with default parameters to clarify the phylogeny of Sinosuthora species. Phylogenetic analysis strongly supported that S. conspicillata is closely related to S. webbiana (). This mitochondrial genome sequence will provide useful genetic information for further studies on the genetic diversity and conservation of the Sylviidae.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are openly available in the US National Center for Biotechnology Information (NCBI), with the reference number of MW393772. https://www.ncbi.nlm.nih.gov/nuccore/MW393772.1/
Additional information
Funding
References
- MacKinnon J, Phillipps K, He FQ. 2000. A field guide to the birds of China. Changsha: Hunan Education Press.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Burland TG. 2000. DNASTAR’s Lasergene sequence analysis software. Methods Mol Biol. 132:71–91.
- Chen P, Sun C, Liu B, Lu C, Chen Y. 2020. Complete mitochondrial genome of the reed parrotbill Paradoxornis heudei (Aves: Passeriformes: Muscicapidae). Mitochondrial DNA Part B. 5(2):1467–1468.
- He W, Xu H, Li D, Xie M, Zhang M, Ni Q, Yao Y. 2019. Complete mitochondrial genome sequence of grey-headed parrotbill (Paradoxornis gularis) and its phylogenetic analysis. Mitochondrial DNA Part B. 4(2):3669–3670.
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19(9):1639–1645.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Wen L, Yang X, Liao J, Fu Y, Dai B. 2017. The complete mitochondrial genome of the fulvous parrotbill Paradoxornis fulvifrons (Passeriformes: Muscicapidae). Mitochondrial DNA. 28(1):143–144.
- Zhang H, Li Y, Wu X, Xue H, Yan P, Wu X. 2015. The complete mitochondrial genome of Paradoxornis webbianus (Passeriformes, Muscicapidae). Mitochondrial DNA. 26(6):879–880.