Abstract
We present the complete mitochondrial genome of Toxotes chatareus yielded by the next-generation sequencing data in this study. The complete mitochondrial genome of T. chatareus has 16,543 bp and contained 13 protein-coding genes, 22 transfer RNA genes, two ribosomal RNA genes, and a single control region (D-loop). The overall base composition was A 28.75%, C 29.80%, G 15.77%, T 25.68% and its gene arrangement was similar with other Carangaria mitochondrial genomes. Additionally, the phylogenetic relationships of 13 Carangaria species based on the complete mitochondrial genome was analyzed using the neighbor-joining method. The result showed T. chatareus was clustered with L. lactarius suggesting the close phylogenetic affinity they owned. Together, the complete mitochondrial genome of T. chatareus would be beneficial for the study of phylogenetic relationship, taxonomic classification and phylogeography of the Carangaria.
Toxotes chatareus belongs to Toxotes; Toxotidae; Carangaria (Allen Citation1978, Citation2004; Simon et al. Citation2010; Schoch et al. Citation2020; Froese and Pauly Citation2021), is mainly distributed in the brackish water of mangrove-lined estuaries along the South Pacific and Indian Oceans, although it can also be found far upstream in fresh waters and more saline coastal waters (Allen Citation2001; Temple Citation2007; Temple et al. Citation2010). It is well known for its ability to shoot down insects with a jet of water squirted from the mouth (Elshoud and Koomen Citation1985; Simon et al. Citation2009, Citation2012). Several researches have focused on the mechanism of spitting (Timmermans Citation2001), visual capabilities (Temple et al. Citation2013) and brain atlas (Karoubi et al. Citation2016). Yet, little information about its genetic characteristics is available. In order to find new DNA markers for the future study of the phylogenetic relationship, taxonomic classification and phylogeography of the Carangaria, we assembled and acquired the complete mitogenome of T. chatareus by the next-generation sequencing (NGS) data.
The T. chatareus sample was acquired from Thailand (14”N; 100”E) and stored in a refrigerator of −80 °C at School of Ecology and Environment, Northwestern Polytechnical University underthe voucher number 20190815AP01. The species was identified based on morphologic features and COI gene (SequenceID: AP006806.1). The NGS data used in this study was produced by our previous study (Lü et al. Citation2021) and could be acquired from the public database NCBI with the accession number SRX8345786. The short reads which were produced from the HiSeq platform were used to assemble the mitogenome by MitoZ software with default parameters (Meng et al. Citation2019). The MITOS Web Server (Bernt et al. Citation2013) and tRNAscan-SE Search Server (Chan & Lowe Citation2019) were used to predict and annotate the mitogenome. Whole mitogenome sequence of T. chatareus was 16,543 bp in length, containing 13 protein-coding genes, 22 tRNA genes, two rRNA genes, and one control region (D-loop). The gene arrangement and base content were similar with other Carangaria species (Lv et al. Citation2016; Wang et al. Citation2016; Gan et al. Citation2017; Yang et al. Citation2019; Tabassum et al. Citation2020). Almost all the protein-coding genes were encoded by H-strand with the exception of ND6 and eight tRNAs (Gln, Ala, Asn, Cys, Tyr, Ser, Glu, Pro) genes located on L-strand. The base composition was A 28.75%, C 29.80%, G 15.77%, T 25.68%. AT and GC contents were 54.43% and 45.57%, respectively. Twelve protein-coding genes started with an ATG initiation codon, while COX1 used GTG as an initiation codon. For the termination codon, eight protein-coding genes (ATP6, ATP8, COX1, COX3, ND1, ND2, ND4L, and ND5) ended with TAA, two protein-coding genes (ND3 and ND6) with TAG, and COX2, CYTB and ND4 have an incomplete stop codon T–. The 13 protein-coding genes were 11,433 bp in length, accounting for 69.11% of the whole mitogenome, which encodes 3,811 amino acids in total. The lengths of 12S rRNA located between tRNAPhe and tRNAVal and 16S rRNA located between tRNAVal and tRNALeu were 959 bp and 1,697 bp, respectively. The control region (D-Loop) typically located between tRNAPro and tRNAPhe, was 828 bp in length.
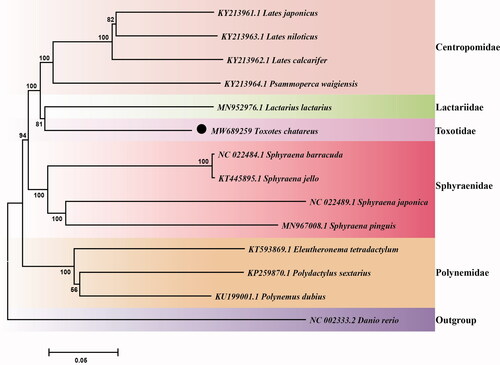
To further investigate the phylogenetic location of T. chatareus in Carangaria, the phylogenetic relationship was constructed in this study. Specifically, we first downloaded the mitochondrial genome of 13 species (including 12 Carangaria species and Danio rerio) in NCBI database. Then, the sequence alignment of these 14 species (including T. chatareus) was conducted by multiple sequence alignment program ClustalW (Thompson et al. Citation1994) in BioEdit software (Hall Citation1999). At last, the phylogenetic tree was constructed using the neighbor joining (NJ) method with 10,000 bootstrap replications using MEGA7 (Kumar et al. Citation2016). The result of the phylogeny shows that T. chatareus is clustered with the L. lactarius (), which is belongs to Lactariidae, suggesting close phylogenetic relationship they owned. We expect that the information of the complete mitogenome of T. chatareus would be beneficial for the study of phylogenetic relationship, taxonomic classification and phylogeography of the Carangaria in the future.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The mitochondrial genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession number MW689259. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA592745, SRR11793814, and SAMN13439313, respectively.
Additional information
Funding
References
- Allen GR. 1978. A review of the archerfishes (family Toxotidae). Rec West Aust Mus. 6.
- Allen GR. 2001. Toxotidae-archer fishes. In: Carpenter KE, Niem VH, editors. FAO species identification guide for fishery purposes. The living marine resources of the Western Central Pacific. Rome: FAO; p. 3212–3215.
- Allen GR. 2004. Toxotes kimberleyensis, a new species of archerfish (Pisces: Toxotidae) from fresh waters of Western Australia. Rec Aust Mus. 56(2):225–230.
- Bernt M, Donath A, J¨Uhling F, Externbrink F, Florentz C, Fritzsch G, P¨Utz J, Middendorf M, Stadler P. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Chan PP, Lowe TM. 2019. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol Biol. 1962:1–14.
- Elshoud GCA, Koomen P. 1985. A biomechanical analysis of spitting in archer fishes (Pisces, Perciformes, Toxidae). Zoomorphology. 105(4):240–252.
- Froese R, Pauly D. 2021. World Wide Web electronic publication. FishBase, www.fishbase.org.
- Gan H, Takahashi H, Hammer M, Tan M, Lee Y, Voss J, Austin C. 2017. Mitochondrial genomes and phylogenetic relationships of Lates japonicus, Lates niloticus, and Psammoperca waigiensis (Perciformes: Latidae). Mitochondrial DNA B Resour. 2(1):73–75.
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 41:95–98.
- Karoubi N, Segev R, Wullimann M. 2016. The brain of the archerfish Toxotes chatareus: a nissl-based neuroanatomical atlas and catecholaminergic/cholinergic systems. Front Neuroanat. 10:106.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Lü Z, Gong L, Ren Y, Chen Y, Wang Z, Liu L, Li H, Chen X, Li Z, Luo H, et al. 2021. Large-scale sequencing of flatfish genomes provides insights into the polyphyletic origin of their specialized body plan. Nat Genet. 53(5):742–751.
- Lv H, Cheng Q, Pang J, Zhang H. 2016. The complete mitochondrial genome sequence of Sphyraena jello (Perciformes: Sphyraenidae) and its phylogenetic position. Mitochondrial DNA A DNA Mapp Seq Anal. 27(6):4570–4571.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Schoch CL, Ciufo S, Domrachev M, Hotton CL, Kannan S, Khovanskaya R, Leipe D, Mcveigh R, O’Neill K, Robbertse B, et al. 2020. NCBI Taxonomy: a comprehensive update on curation, resources and tools. Database (Oxford). 2020.
- Simon KD, Bakar Y, Mazlan AG, Zaidi CC, Samat A, Arshad A, Temple SE, Brown-Peterson NJ. 2012. Aspects of the reproductive biology of two archer fishes Toxotes chatareus, (Hamilton 1822) and Toxotes jaculatrix (Pallas 1767). Environ Biol Fish. 93(4):491–503.
- Simon KD, Bakar Y, Samat A, Zaidi CC, Aziz A, Mazlan AG. 2009. Population growth, trophic level, and reproductive biology of two congeneric archer fishes (Toxotes chatareus, Hamilton 1822 and Toxotes jaculatrix, Pallas 1767) inhabiting Malaysian coastal waters. J Zhejiang Univ Sci B. 10(12):902–911.
- Simon K, Bakar Y, Temple S, Mazlan A. 2010. Morphometric and meristic variation in two congeneric archer fishes Toxotes chatareus (Hamilton 1822) and Toxotes jaculatrix (Pallas 1767) inhabiting Malaysian coastal waters. J Zhejiang Univ Sci B. 11(11):871–879.
- Tabassum N, Park W, Baek H, Je J, Kim H. 2020. Characterization of the complete mitochondrial genome of brown barracuda, Sphyraena pinguis (Perciformes: Sphyraenidae). Mitochondrial DNA B Resour. 5(3):3042–3043.
- Temple SE. 2007. Effect of salinity on the refractive index of water: considerations for archer fish aerial vision. Journal of Fish Biology. 70(5):1626–1629.
- Temple S, Hart NS, Marshall NJ, Collin SP. 2010. A spitting image: specializations in archerfish eyes for vision at the interface between air and water. Proc Biol Sci. 277:2607–2615.
- Temple SE, Manietta D, Fau-Collin SP, Collin SP. 2013. A comparison of behavioural (Landolt C) and anatomical estimates of visual acuity in archerfish (Toxotes chatareus). Vision Res. 83:1–8.
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22(22):4673–4680.
- Timmermans P. 2001. Prey catching in the archer fish: angles and probability of hitting an aerial target. Behav Processes. 55(2):93–105.
- Wang C, Qiu J, Peng X, Ai W, Huang X, Liu W, Chen S. 2016. The complete mitochondrial genome of Polydactylus sextarius (Teleostei, Mugiliformes). Mitochondrial DNA A DNA Mapp Seq Anal. 27(5):3344–3345.
- Yang M, Li P, Qin Q, Zhu K. 2019. The complete mitochondrial genome of false trevally Lactarius Lactarius (Bloch and Schneider, 1801). Mitochondrial DNA B Resour. 5(1):87–89.