Abstract
In the study, the complete mitochondrial genome of Artemia salina was reported for the first time. The mitochondrial genome of A. salina is 15,762 bp in length, with the typical structure of 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), and two ribosomal RNA genes, and a major non-coding region (CR). Phylogenetic analysis showed that A. salina has a much closer relationship with A. persimilis compared to other Artemia species. The complete cp genome sequence of A. salina reported here provided an essential resource for further population genetics research and germplasm conservation on Artemia.
Brine shrimp Artemia (Arthropoda, Crustacea, Anostraca) inhabits hypersaline environments such as salt lakes and solar saltworks, which plays a biological regulatory role in salt field ecosystem. The genus Artemia is generally considered to contain seven bisexual species as well as some parthenogenetic Artemia populations with different polyploidy types (Asem et al. Citation2010). At present, the complete mitochondrial genome has been reported in four bisexual species, including Artemia franciscana, Artemia urmiana, Artemia tibetiana, and Artemia sinica (Valverde et al. Citation1994; Zhang et al. Citation2013; Asem et al. Citation2019). As for Artemia salina, a bisexual species endemic to the Mediterranean Basinare, is being threatened by habitat loss and the ongoing A. franciscana invasion (Muñoz et al. Citation2008). However, its complete mitochondrial genome has not been reported and characterized. Herein, we characterized the complete mitochondrial genome sequence of A. salina using Illumina sequencing, and a phylogenetic analysis was performed to investigate the phylogenetic relationships of A. salina with other Artemia species.
The cysts of A. salina were collected from Sebkha Eladhibet Saltworks, Tunisia (**latitude** 11.4342 and **longitude** 33.1060). The specimen was deposited at the Asian Regional Artemia Reference Center (Tianjin University of Science and Technology, Tianjin, China) (Liying Sui, [email protected]) under the voucher number 1632. The genomic DNA was extracted form cysts using TIANGEN®TIANamp Genomic DNA Kit (Tianjin, China). Then, the high quality gDNA was sequenced using Illumina Novaseq6000 platform with 350 bp insert size. The complete mitochondrial genome was assembled using SPAdes v.3.5.0 (http://cab.spbu.ru/software/spades/) with A. franciscana (GenBank accession number: X69067) as reference. The genome was first annotated with MITOS (http://mitos.bioinf.uni-leipzig.de/index.py) and ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/), then the reference mitochondrial map and BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) were used to verify the accuracy of the results and make corrections. The tRNA genes were predicted using the ARWEN (http://mbio-serv2.mbioekol.lu.se/ARWEN/) and tRNAscan-SE 2.0 (http://lowelab.ucsc.edu/tRNAscan-SE/) online software.
The complete mitochondrial genome of A. salina (GenBank accession number: MZ199177) is 15,762 bp in length, with the typical structure of 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), and two ribosomal RNA genes, and a major non-coding region (CR) called the D-loop. The length variations are mainly confined to the D-loop region. The base composition is 30.80% A, 17.49% C, 17.98% G, and 33.73% T, with an A + T content of 64.53%. Just five PCGs (cox1, atp6, cox3, cytb, and nd1) began with the common ATG start codon. Stop codons included TAA (cox3, nd2, atp8, atp6, nd3, and cytb) and TAG (nd1, nd4l, and nd6). There are four genes ended with the incomplete stop codon T (cox1, cox2, nd4, and nd5). The 12S rRNA and16S rRNA were separated by the trnV. The length of rrnL and rrnS is 1144 bp and 711 bp, respectively. The 22 tRNA genes size varies from 59 to 67 bp, respectively.
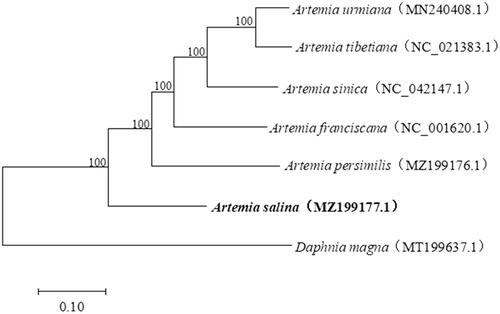
To further explore the phylogenetic relationships among Artemia species, the complete mitochondrial genome sequences of five bisexual Artemia species were downloaded from GenBank, and the software MEGA 7.0 was used to make phylogenetic analysis by maximum-likelihood (ML) method with the Kimura 2-parameter model (Kumar et al. Citation2016). The results showed that A. salina at the base position of Artemia in this tree, and A. salina has a much closer relationship with A. persimilis compared to other Artemia species (). The mitochondrial genome sequence of A. salina reported here can provide important information for population genetics and evolutionary studies of Artemia.
Acknowledgements
The authors thank Artemia Reference Center (Ghent University, Belgium) for assistance with specimen collection.
Disclosure statement
The authors declare that they have no conflict of interest.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession number MZ199177. The associated BioProject, SRA, and BioSample numbers are PRJNA749918, SRR15292986, and SAMN20425675, respectively.
Additional information
Funding
References
- Asem A, Rastegar-Pouyani N, De los Rios P. 2010. The genus Artemia Leach, 1819 (Crustacea: Branchiopoda): true and false taxonomical descriptions. Lat Am J Aquat Res. 38:501–506.
- Asem A, Li WD, Pei-Zheng Wang PZ, Eimanifar A, Shen CY, De Vos S, Van Stappen G. 2019. The complete mitochondrial genome of Artemia sinica Cai, 1989 (Crustacea: Anostraca) using next-generation sequencing. Mitochondrial DNA B. 4(1):746–747.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Bio Evol. 33(7):1870–1874.
- Muñoz J, Gómez A, Green AJ, Figuerola J, Amat F, Rico C. 2008. Phylogeography and local endemism of the native Mediterranean brine shrimp Artemia salina (Branchiopoda: Anostraca). Mol Ecol. 17(13):3160–3177.
- Valverde J, Batuecas B, Moratilla C, Marco R, Garesse R. 1994. The complete mitochondrial DNA sequence of the Crustacean Artemia franciscana. J Mol Evol. 39(4):400–408.
- Zhang HX, Luo QB, Sun J, Liu F, Wu G, Yu J, Wang WW. 2013. Mitochondrial genome sequences of Artemia tibetiana and Artemia urmiana: assessing molecular changes for high plateau adaptation. Sci China Life Sci. 56(5):440–452.