Abstract
Amaranthus dubius is a leafy vegetable widely cultivated in Asia and Africa. The complete chloroplast genome of Amaranthus dubius was sequenced and assembled in this study. The complete chloroplast genome is 150,520 bp. A total of 130 genes were identified, including 85 protein-coding genes, eight rRNA genes, and 37 tRNA genes. The overall GC content of this genome was 36.6%. The phylogenetic tree based on 10 chloroplast genomes in Amaranthaceae supports that A. dubius is sister to A. hypochondriacus and A. caudatus.
Amaranthus dubius Mart. ex Thell. 1912 (Amaranthaceae), an annual herb native to tropical America, is widely cultivated as a green vegetable and considered a medicinal herb in many Asian and African countries (Achigan-Dako et al. Citation2014; Alegbejo Citation2014). Studies have shown that the leaves of A. dubius can be used as complement dietary for rice, wheat, and corn proteins (Rodríguez et al. Citation2011). For the past decades, it has escaped from cultivation and now it is considered naturalized throughout the tropical and subtropical regions of Europe, Asia, and Africa (Wang et al. Citation2015). Despite being used as an important vegetable, little molecular genetic information of this species has been reported. A recent study showed that there is a disagreement between the concatenated chloroplast genes and nuclear datasets in the placement of the species, which indicates a possible complex evolutionary history of this species (Waselkov et al. Citation2018). Additionally, A. dubius is the only known allotetraploid Amaranthus species. To further study its species origin, we assembled and reported the complete chloroplast genome of A. dubius here for the first time. We hope it will provide valuable genetic resources for comparative genomic studies in resolving the evolution and underlying genetic questions in Amaranthus.
The A. dubius individual was collected from Fuzhou, China (GPS: E 116°20′25.08″, N 26°50′37.32″). DNA was extracted from its dried specimen leaves using Plant Genomic DNA Kit (Tiangen, Beijing, Co., Ltd., Beijing, China). The specimen and extracted DNA was deposited at Shanghai Chenshan Herbarium (CSH), Shanghai Chenshan Botanical Garden (http://www.csnbgsh.cn) under the voucher number RQHD03140 (collected by Xiao-Ling Yan: [email protected]). The plastome sequences were generated using the Illumina HiSeq 2500 platform (Illumina Inc., San Diego, CA). In total, about 16 million high-quality clean reads (150 bp PE read length) were generated with adaptors trimmed. Aligning, assembly, and annotation were conducted by GetOrganelle (Jin et al, Citation2020), GeSeq (Tillich et al. Citation2017), and GENEIOUS v11.1.5 (Biomatters Ltd., Auckland, New Zealand).
The full length of A. dubius chloroplast sequence (GenBank accession no. MZ397802) is 150,520 bp. It is comprised by a large single copy region (LSC with 83,869 bp), a small single copy region (SSC with 17,947 bp), and two inverted repeat regions (IR with 24,352 bp). The GC content of A. dubius chloroplast genome was 36.6% and the GC contents of the LSC, SSC, and IR regions are 34.5%, 30.3%, and 42.6%. The genome (85 protein-coding genes, eight rRNA genes, and 37 tRNA genes) contained 130 genes. Seventeen genes had two copies, which were comprised of six PCG genes (ndhB, rpl2, rpl23, rps12, rps7, ycf2), seven tRNA genes (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, trnV-GAC), and all four rRNA species (rrn16, rrn23, rrn4.5, rrn5). In the genome, seven protein-coding genes (rps16, atpF, rpoC1, petB, rpl16, ndhB, ndhA) had one intron, and three protein-coding genes (rps12, ycf3, clpP) contained two introns.
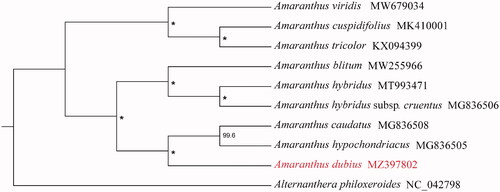
To analyze the phylogenetic placement of A. dubius in Amaranthus, chloroplast genomes sequences of additional eight Amaranthus species and Alternanthera philoxeroides as outgroup were obtained from NCBI. The sequence alignment was conducted by MAFFT v7.450 (Katoh and Standley Citation2013), of which parameter used the default. Based on TVM + F+G4 model and 5000 bootstrap replicates, the maximum-likelihood (ML) analysis was performed by using IQTREE v2.0.6 (Nguyen et al. Citation2015). The result indicates that A. dubius is sister to a clade formed by A. hypochondriacus and A. caudatus (). We hope the complete chloroplast genome of A. dubius will provide necessary genetic resource and background data for further phylogenetic study of the Amaranthaceae.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov) under the accession no. MZ397802. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA741594, SRR14920259, and SAMN19882390, respectively. The DNA matrix and phylogenetic tree that support the findings of this study are openly available on figshare: https://doi.org/10.6084/m9.figshare.16530771.
Additional information
Funding
References
- Achigan-Dako EG, Sogbohossou OE, Maundu P. 2014. Current knowledge on Amaranthus spp.: research avenues for improved nutritional value and yield in leafy amaranths in sub-Saharan Africa. Euphytica. 197(3):303–317.
- Alegbejo JO. 2014. Nutritional value and utilization of Amaranthus (Amaranthus spp.) – a review. Bayero J Pure Appl Sci. 6(1):136–143.
- Jin JJ, Yu WB, Yang JB, Song Y, DePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):1–31.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Rodríguez P, Pérez E, Romel G, Dufour D. 2011. Characterization of the proteins fractions extracted from leaves of Amaranthus dubius (Amaranthus spp.). Afr J Food Sci. 5(7):417–424.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq- versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wang Q, Wang Y, Yan X, Zeng X, Ma J, Li H. 2015. Amaranthus dubius Mart. ex Thell.—a newly naturalized plant of mainland of China. J Trop Subtrop Bot. 23(3):284–288.
- Waselkov KE, Boleda AS, Olsen KM. 2018. A phylogeny of the genus Amaranthus (Amaranthaceae) based on several low-copy nuclear loci and chloroplast regions. Syst Bot. 43(2):439–458.