Abstract
In the present study, we reported and characterized the complete chloroplast genome of a moth orchid, Phalaenopsis wilsonii, which is endemic to South China. Its plastid genome size is 145,373 bp, consisting of a large single copy (LSC) region (84,996 bp), a small single-copy region (10,668 bp), and two inverted repeats (IRs) regions (24,855 bp). A total of 122 plastid genes were annotated, comprising 76 protein-coding genes, 38 tRNA genes, and 8 rRNA genes. The phylogenetic tree further revealed that P. wilsonii showed a sister relationship with P. lowii within subgenus Parishianae.
Moth orchid (Phalaenopsis Blume) is widely used in gardening around the world, and also occupies a large proportion of orchid industry (Van Huylenbroeck Citation2018). Previously phylogenetic analyses revealed that Phalaenopsis can be divided into four subgenera, subgen. Phalaenopsis, Parishianae, Hygrochilus and Ornithochilus (Kocyan and Schuiteman Citation2014; Li et al. Citation2014, Citation2016). Although Subgen. Parishianae constitutes more than a half of the species richness in Phalaenopsis, only one complete chloroplast genome of this subgenus can be found in NCBI (Wang et al. Citation2019). P. wilsonii is an endemic species in China, and is also a typical deciduous Phalaenopsis belonging to subgen. Parishianae. Here, we provide a complete chloroplast genome of P. wilsonii, aiming to facilitate our understanding on Phalaenopsis as well as to expand orchid resources in wild.
The sampling individual of Phalaenopsis wilsonii is cultivated in National Orchid Conservation Center in Guangdong province of China (114°19’01′’E, 22°60’34′’N), and a voucher specimen (noccphal031n) was also deposited in the Herbarium of National Orchid Conservation Center, Shenzhen, China. We extracted the total DNA from the young leaf of the voucher specimen and conducted high throughput sequencing at Illumina HiSeq 2000 platform (Illumina, San Diego, CA). Firstly, we constructed a reference dataset with all publicly available Phalaenopsis plastid genomes, and then mapped the clean reads against the reference dataset to obtain the chloroplast reads for P. wilsonii. PLATANUS (Kajitani et al. Citation2014) was adopted for contig assembly and scaffolding, resulting in the final complete genome with the artificial modification. BLAST was further used to align the reads onto the genome again to determine the IR boundaries, and the annotation was performed using Geneious 2019.0.3 (Kearse et al. Citation2012). The resulting complete chloroplast genome of P. wilsonii was submitted to GenBank under the accession number of MW218959. The total length of P. wilsonii chloroplast genome is 145,373 bp, with the GC content being 36.9%. The chloroplast genome we identified here is slightly shorter than other published moth orchids (from 146,834 bp for P. lowii to 148,964 bp for P. aphrodite subsp. formosana). As with other orchids, the chloroplast genome of P. wilsonii consists of a large single copy (LSC) region (84,995 bp) and a small single-copy region (10,668 bp), which are segmented by two inverted repeat (IRs) regions (24,855 bp). Overall, 122 genes (containing repeat region gene) were annotated, including 76 protein-coding genes, 8 rRNAs, and 38 tRNAs. Similar to a former study (Chang et al. Citation2006), all ndh genes of P. wilsonii are nonfunctional, and the ndhE was also missing.
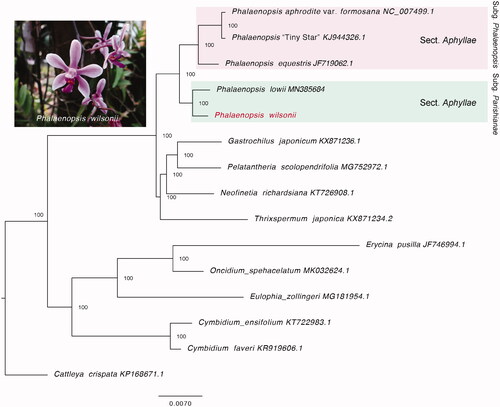
To infer the phylogenetic relationship for P. wilsonii, we used RAxML v.8 (Stamatakis Citation2014) to reconstruct the maximum-likelihood phylogenetic tree based on the whole plastid genomes of P. wilsonii, four moth orchids, and also the other ten orchids. Cattleya crispate was applied as an outgroup according to the topology from Givnish et al. (Citation2015). The TIM1 + I + G model was applied by jModelTest (Posada Citation2008), and the reliability of topology was supported with 1000 bootstrap replicates. Consistent with Givnish’s study Givnish et al. (Citation2015), the phylogenetic tree showed that Vandeae presented a sister relationship to Cymbidieae. Besides, P. wilsonii was sister to P. lowii, both of which belong to the subgen. Parishianae that are grouped into a single clade (). This complete chloroplast genome of P. wilsonii will be helpful for future phylogenetic studies and conservation in Phalaenopsis.
Authors’ contributions
J.Y.W. and K.K.X. conceived the study; J.Y.W. and K.K.X. obtained the molecular data; J.Y.W. and D.K.L. conducted the data analysis; K.K.X. drafted the manuscript; J.Y.W. and D.K.L. revised the manuscript. All authors provided comments and final approval.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that newly obtained at this study are available in the NCBI under accession number of MW218959 (https://www.ncbi.nlm.nih.gov/nuccore/MW218959). The sequencing reads are available under the SRA accession number of SAMN18924765.
Additional information
Funding
References
- Chang C-C, Lin H-C, Lin I-P, Chow T-Y, Chen H-H, Chen W-H, Cheng C-H, Lin C-Y, Liu S-M, Chang C-C, et al. 2006. The chloroplast genome of Phalaenopsis aphrodite (Orchidaceae): comparative analysis of evolutionary rate with that of grasses and its phylogenetic implications. Mol Biol Evol. 23 (2):279–291.
- Givnish TJ, Spalink D, Ames M, Lyon SP, Hunter SJ, Zuluaga A, Iles WJD, Clements MA, Arroyo MTK, Leebens-Mack J, et al. 2015. Orchid phylogenomics and multiple drivers of their extraordinary diversification. Proc R Soc B. 282(1814):20151553.
- Kajitani R, Toshimoto K, Noguchi H, Toyoda A, Ogura Y, Okuno M, Yabana M, Harada M, Nagayasu E, Maruyama H, et al. 2014. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 24(8):1384–1395.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kocyan A, Schuiteman A. 2014. New combinations in Aeridinae (Orchidaceae). Phytotaxa. 161 (1):61–85.
- Li M-H, Gruss O, Liu Z-J. 2016. Nomenclature changes in Phalaenopsis subgen. Hygrochilus (Orchidaceae; Epidendroideae; Vandeae) based on DNA evidence. Phytotaxa. 275(1):55–61.
- Li M-H, Zhang G-Q, Liu Z-J, Lan S-R. 2014. Revision of Hygrochilus (Orchidaceae: Epidendroideae: Aeridinae) and a molecular phylogenetic analysis. Phytotaxa. 159(4):256.
- Posada D. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol. 25(7):1253–1256.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Van Huylenbroeck J. 2018. Ornamental crops. Springer.
- Wang J-Y, Liu Z-J, Zhang G-Q, Peng C-C. 2019. The complete chloroplast genome sequence of Phalaenopsis lowii (Orchidaceae). Mitochondrial DNA B Resour. 4(2):3569–3570.