Abstract
Impatiens mengtszeana is an endemic species in China. In this study, the complete chloroplast genome of I. mengtszeana was sequenced and analyzed. The total chloroplast genome size of I. mengtszeana is 152,928 bp, including a pair of inverted repeat regions (IRs, 26,007 bp) separated by a large single copy (LSC, 83,722 bp) region and a small single copy region (SSC, 17,192 bp). The whole chloroplast genome contains 89 protein-coding genes (PCGs), 37 transfer RNA genes (tRNAs), and eight ribosomal RNA genes (rRNAs). According to the phylogenetic topologies, I. mengtszeana was closely related to I. hawkeri.
Impatiens is a large genus in the family Balsaminaceae, comprises over 1000 species, mainly distributed in tropical and subtropical regions (Yu et al. Citation2016; Stefaniak et al. Citation2020). It is known for its multi-colors flowers and ornamental values. The chemical substances in the rhizome of Impatiens are widely used in the field of medicine (Zhou et al. Citation2015; Chua Citation2016). Impatiens is considered to be one of the most taxonomically difficult groups in angiosperm. The species within the genus are extremely variable and lack the synapomorphic characters necessary to define subdivisions. (Grey-Wilson Citation1980; Yu Citation2012). Impatiens mengtszeana is an annual herb, endemic to south-west China (Lan et al. Citation2004; Yu Citation2012). Flowers are light yellow with red streak. At present, there are few studies on I. mengtszeana. Herein, it is necessary to describe evolutionary relationships in terms of relative species of Impatiens based on highthroughput sequencing approaches. This study provides important information of Balsaminaceae on evolution, genetic and molecular biology for future studies.
The fresh leaves of I. mengtszeana were sampled from Malipo County, Wenshan City, Yunnan Province, China (104°50′39.73″ E, 23°10′9.07″ N, 1420 m altitude) and DNA samples were stored in Guizhou Normal University (accession number: TR-010). Total DNA was extracted from fresh leaves by CTAB method and applied to 500-bp paired-end library construction using the TruSeq Nano DNA Sample Prep Kit for Illumina sequencing. Sequencing was carried out on the Illumina NovaSeq 6000 platform (BIOZERON Co., Ltd, Shanghai, China). Approximately 2.4 Gb of raw data from I. mengtszeana were generated with 150 bp paired-end read lengths. The low quality sequences of raw reads used Trimmomatic 0.39 for quality control (Bolger et al. Citation2014). The trimmed reads were selected for cp genome assembly by NOVOPlasty4.2 software (Dierckxsens et al. Citation2017). The complete chloroplast genome of I. mengtszeana was annotated by GeSeq (Tillich et al. Citation2017). For a better understanding of I. mengtszeana, the physical map of the new chloroplast genome was generated using OGDRAW (Lohse et al. Citation2013). The complete cp genome was submitted to Genbank (accession number of MW727522).
The complete cp genome sequence of I. mengtszeana is 152,928 bp and shows a characteristic circular structure. Overall GC content of the whole genome is 36.73%. A large single-copy (LSC, GC-34.34%) region of 83,722 bp and a small single-copy (SSC, GC-29.53%) region of 17,192 bp, which is separated by a pair of inverted repeat (IRa and IRb) regions of 26,007 bp. There are a total of 89 protein-coding genes, 37 tRNA genes, and eight rRNA genes. Among these, duplication events occurred with 19 genes (rps12, rp12, rp123, ycf2, ycf15, ndhB, rps7, ycf1, trnA-UGC, trnG-GCC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnV-GAC, rrn16, rrn23, rrn4.5, rrn5).
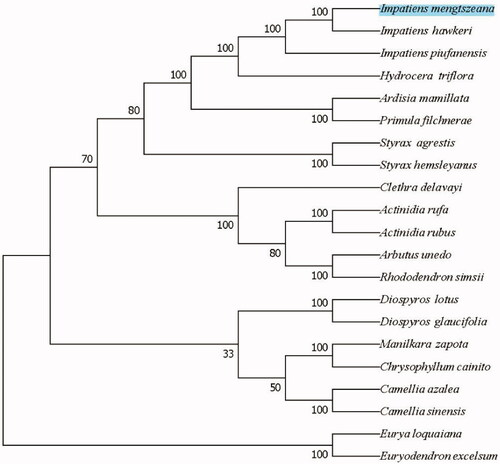
In order to further determine the phylogenetic location of I. mengtszeana, 20 cp genome sequences from Ericales were downloaded from GenBank. BioAider1.0 software was used for the alignment of cp genome sequences (Zhou et al. Citation2020). A neighbor-joining phylogenetic tree was constructed in MEGA7 software with 1000 bootstrap replicates (Kumar et al. Citation2016). The NJ tree analysis indicated that I. mengtszeana is closely related to Impatiens hawkeri with 100% bootstrap support ().
Figure 1. The NJ phylogenetic tree for I. mengtszeana based on 20 chloroplast genome sequences of Ericales. Numbers on the nodes are bootstrap values from 1000 replicates. Accession numbers: Actinidia rufa (NC_039973.1), Actinidia rubus (MN652056.1), Clethra delavayi (NC_041129.1), Impatiens hawkeri (NC_048520.1), Impatiens piufanensis (NC_037401.1), Hydrocera triflora (NC_037400.1), Diospyros lotus (NC_030786.1), Diospyros glaucifolia (NC_030784.1), Arbutus unedo (JQ067650.2), Rhododendron simsii (MW030509.1), Eurya loquaiana (NC_050937.1), Euryodendron excelsum (NC_039178.1), Primula filchnerae (NC_051972.1), Ardisia mamillata (MN136062.1), Manilkara zapota (MN295595.1), Styrax agrestis (MT644192.1) and Styrax hemsleyanus (NC_047298.1).
Disclosure statement
The authors report no conflict of interest and are responsible for the content.
Data availability statement
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/nuccore/ MW727522, reference number MW727522.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Chua LS. 2016. Untargeted MS-based small metabolite identification from the plant leaves and stems of Impatiens balsamina. Plant Physiol Biochem. 106:16–22.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Grey-Wilson C. 1980. Impatiens of Africa. Rotterdam: Belkema Press.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Lan Q-Y, Yin S-H, He H-Y. 2004. In vitro conservation of Impatiens mengtzeana. Acta Botanica Boreali-Occidentalia Sinica. 24(1):146.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41(Web Server issue):W575–W581.
- Stefaniak AR, Adamowski W, Borah S, Gogoi R. 2020. New data on seed coat micromorphology of several Impatiens spp. from northeast India. Acta Soc Bot Pol. 89(3):89312.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Yu S-X, Janssens SB, Zhu X-Y, Liden M, Gao T-G, Wang W. 2016. Phylogeny of Impatiens (balsaminaceae): integrating molecular and morphological evidence into a new classification. Cladistics. 32(2):179–197.
- Yu S-X. 2012. Balsaminaceae of China. Beijing: Peking University Press.
- Zhou X-R, Wang H-X, Li J-H, Mao L. 2015. Situation and prospects of floral resources in Balsaminiaceae. Hubei Agric Sci. 02:266–269.
- Zhou Z-J, Qiu Y, Pu Y, Huang X, Ge X-Y. 2020. BioAider: an efficient tool for viral genome analysis and its application in tracing SARS-CoV-2 transmission. Sustain Cities Soc. 63:102466.
