Abstract
Crataegus bretschneideri Schneid., with an unclear phylogenic position, is mainly distributed in northeast and inner mongolia area of China. In this study, the complete chloroplast genome sequence of C. bretschneideri was determined by using Illumina high-throughput sequencing method. The chloroplast genome was 159,607 bp in length and consisted of a large single-copy (LSC) region (87,601 bp), a small single-copy (SSC) region (19,312 bp), separated by a pair of inverted repeat (IRs: 26,347 bp, each) regions. It comprised a total of 114 unique genes, including 80 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. Phylogenetic analysis based on complete chloroplast genomes indicated that C. bretschneideri was closely related to C. marshallii Eggl in the subfamily Maloideae. This complete chloroplast genome will provide valuable insight into evolution, molecular breeding, and phylogenetic analysis of Crataegus species.
The genus Crataegus (hawthorn), a member of the Rosaceae family, is widely distributed throughout temperate regions in the Northern Hemisphere including Eurasia and North America (Phipps et al. Citation1990; Christensen Citation1992; Du et al. Citation2019). Hawthorns are one of the most important processing and table fruits in China, owing to their nutrient-rich fruit and significant medicinal values (Özcan et al. Citation2005; Xu et al. Citation2016; Zheng et al. Citation2018). A total of 18 species and six varieties of Crataegus have been confirmed in China (Zhao and Feng Citation1996; Xin and Zhang Citation1997). Crataegus bretschneideri Schneid., originated from Changbaishan Massif of China, is mainly distributed in northeast and inner mongolia area of China (Zhao and Feng Citation1996). It is an important germplasm of Crataegus in China, with the characteristics of high yield, early-maturing and cold resistance. Its fruit is rich in nutrition, especially in natural red pigment, and has excellent processing properties.
C. bretschneideri is morphologically very similar to C. pinnatifida Bge., and Dai (Citation2007) consider the former to be a variant of the latter species. Most of the morphological characters, including leaf color, leaf shape, leaf margin, fruit shape, peel color, etc, are similar between the two species. The most obvious differences between C. bretschneideri and C. pinnatifida are in the leaf blade lobes and seed number. Based on peroxidase isozymograms and inter-simple sequence repeat (ISSR) markers, some researchers suggest that C. bretschneideri is closely related to C. pinnatifida (Schneider Citation1906; Guo and Jiao Citation1995; Han et al. Citation2009). Specific locus amplified fragment sequencing revealed that C. bretschneideri was derived from the hybridization of C. pinnatifida with C. maximowiczii Schnieid (Du et al. Citation2019).
Chloroplast genomes are important sources for taxonomic classification and phylogenetic reconstruction of plant species (Dong et al. Citation2017; Liu et al. Citation2020; Wang et al. Citation2021). In order to clarify the phylogenetic position of C. bretschneideri, we reported the complete chloroplast genome based on Illumina sequencing data (GenBank accession number: MW963339), which would be helpful for evolution, phylogenetic analysis and molecular breeding.
The sample of C. bretschneideri was collected from the Hawthorn Germplasm Repository of Beijing Academy of Forestry and Pomology Sciences (39°97′N, 116°23′E) in Beijing, China. A specimen was deposited at the Herbarium of Beijing Academy of Forestry and Pomology Sciences (BAFPSH, http://www.lgs.baafs.net.cn/, Yuanyong Qi, [email protected]) under the voucher number BJLGY-2020-SZ002. The total genomic DNA from leaves was extracted using a modified CTAB method (Li et al. Citation2013) and paired-end libraries were prepared with the NEBNext Ultra DNA Library Prep Kit. High-throughput sequencing was carried out using the HiSeq Xten PE150 System (Illumina, San Diego, CA, USA) with150bp pair-end reads. In all, 1.72 G raw reads were obtained, and after the quality-trimmed using the software CLC Genomics Workbench v7.5 (CLC bio, Aarhus, Denmark), 1.72 G qualified reads were assembled using SPAdes 3.6.1 (Kmer = 95) (Bankevich et al. Citation2012) to contigs. The contigs of chloroplast genome were selected with the BLAST program (Altschul et al. Citation1990), taking the closely related species C. hupehensis (MW201730) as a reference, and the selected contigs were assembled using Sequencher 4.10 (https://www.genecodes.com/) software tools. Annotation was performed using the Plann (Huang and Cronk Citation2015), then a physical map of the chloroplast genome generated by Genome Vx (Conant and Wolfe Citation2008).
The cp genome of C. bretschneideri was 159,607 bp in length, and consisted of a large single-copy (LSC) region (87601 bp), a small single-copy (SSC) region (19312 bp), separated by a pair of inverted repeat (IRs: 26,347 bp, each) regions. The total GC content of complete chloroplast genome, LSC, SSC, IR regions were 36.6%, 34.4%, 30.3% and 42.7%, respectively. The chloroplast DNA of C. bretschneideri comprised a total of 114 unique genes, including 80 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. In these genes, 19 genes were duplicated in the IR regions, 15 genes harbored a single intron, and 2 (ycf3, clpP) contained double introns.
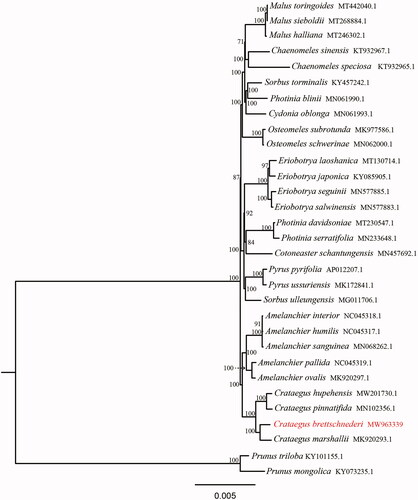
To clarify the phylogenetic position of C. bretschneideri, total 31 complete chloroplast genomes were obtained from Genbank and the sister group Prunoideae was taken as an out group. All chloroplast genome sequences were aligned using MAFFT (Katoh et al. Citation2019), which has been deposited at doi:10.5061/dryad.qv9s4mwfg. Phylogenetic analysis was conducted using maximum-likelihood (ML) method by IQ-TREE (1.6.12) with 1000 bootstrap replicates (Nguyen et al. Citation2015). The phylogenetic analysis showed that C. bretschneideri was closely related to C. marshallii Eggl, rather than C. pinnatifida in the subfamily Maloideae (). This suggests that C. bretschneideri is a distinct Crataegus species, rather than a variant of C. pinnatifida. The phylogenetic tree can provide reference for the parent selection in hawthorn breeding programme. This complete chloroplast genome can be used for future studies on genetic engineering, population and phylogeny of family Rosaceae.
Disclosure statement
No potential conflict of interest was reported by the author(s)
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW963339. The associated BioProject and Bio-Sample numbers are PRJNA722683, and SAMN18789926 respectively.
Additional information
Funding
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Christensen KI. 1992. Revision of Crataegus sect. Crataegus and nothosect. Crataeguineae (Rosaceae-Maloideae) in the Old World. Syst. Bot. Monogr. 35:1–199.
- Conant GC, Wolfe KH. 2008. GenomeVx: simple web-based creation of editable circular chromosome maps. Bioinformatics. 24(6):861–862.
- Dai HY. 2007. Molecular identification and enhancement of germplasms in hawthorn [Ph.D. Thesis]. Shenyang: Shenyang Agricultural University.
- Dong W, Xu C, Li W, Xie X, Lu Y, Liu Y, Jin X, Suo Z. 2017. Phylogenetic resolution in Juglans based on complete chloroplast genomes and nuclear DNA sequences. Front Plant Sci. 8:1148.
- Du X, Zhang X, Bu XD, Zhang TC, Lao YC, Dong WX. 2019. Molecular analysis of evolution and origins of cultivated hawthorn (Crataegus spp.) and related species in China. Front Plant Sci. 10:443.
- Guo TJ, Jiao PJ. 1995. Hawthorn (Crataegus) resources in China. HortSci. 30(6):1132–1134.
- Han XY, Ling YH, Wang YJ, Li F, Guo TJ, Xue YJ. 2009. Analysis of the origin and classification of C. bretschneideri by ISSR Markers. J Jilin Agricultural University. 31(2):164–167.
- Huang DI, Cronk Q. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3(8):1500026.
- Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4):1160–1166.
- Li JL, Wang S, Yu J, Wang L, Zhou SL. 2013. A modified CTAB protocol for plant DNA extraction. Chin Bull Bot. 48(1):72–78.
- Liu Q, Li XY, Li MZ, Xu WK, Schwarzacher T, Heslop-Harrison JS. 2020. Comparative chloroplast genome analyses of Avena: insights into evolutionary dynamics and phylogeny. BMC Plant Biol. 20(1):406.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Özcan M, Hacıseferoğullar H, Marakoğlu T, Arslan D. 2005. Hawthorn (Crataegus spp.) fruit: some physical and chemical properties. J. Food Eng. 69(4):409–413.
- Phipps JB, Robertson KR, Smith PG, Rohrer JR. 1990. A checklist of the subfamily Maloideae (Rosaceae). Can J Bot. 68(10):2209–2332.
- Schneider CK. 1906. Crataegus bretschneideri Schneid. in Repert Spec Nov Regni Veg. 3:223.
- Wang Y, Wang S, Liu Y, Yuan Q, Sun J, Guo L. 2021. Chloroplast genome variation and phylogenetic relationships of Atractylodes species. BMC Genomics. 22(1):103.
- Xin XG, Zhang YM. 1997. Chinese Hawthorn germplasm resources and utilization. Beijing: China Agricultural Press.
- Xu J, Zhao Y, Zhang X, Zhang L, Hou Y, Dong W. 2016. Transcriptome analysis and ultrastructure observation reveal that hawthorn fruit softening is due to cellulose/hemicellulose degradation. Front Plant Sci. 7:1524.
- Zhao HC, Feng BT. 1996. China rruit-plant monograph of hawthorn (Crataegus) Flora. Beijing: ZhongguoLinye Press.
- Zheng GQ, Deng J, Wen LR, You LJ, Zhao ZG, Zhou L. 2018. Release of phenolic compounds and antioxidant capacity of Chinese hawthorn “Crataegus pinnatifida” during, in vitro, digestion. J. Funct. Foods. 40:6–85.