Abstract
The pink sea fan, Eunicella verrucosa (Pallas, 1766), inhabits rocky substrates across the northeast Atlantic and the western Mediterranean. Across much of its range it has been detrimentally affected by fishing. DNA from 17 E. verrucosa specimens was amplified by phi29-induced rolling circle amplification. Following purification by sodium acetate-ethanol precipitation, the circular genomic DNA was sequenced on an Illumina MiSeq v2. Specimens originated from sites along the west coast of Ireland, southwest Wales, southwest/southern England, northwest France, southern Portugal, and the Mediterranean coast of northeast Spain. All samples had identical mitochondrial genome sequences of 19,267 bp and included 14 protein-coding genes (including the mutS gene), two ribosomal RNA subunits (12S and 16S) and one methionine tRNA gene. Two genes (nad2 and nad5) overlapped by 13 bp; all other genes were separated by non-coding intergenic regions. All protein-coding genes had the same start codon (ATG) and a TAA or TAG stop codon, except for cox1 that terminated with the incomplete stop codon T--. The mitochondrial genome of E. verrucosa (MW588805) showed 99.72% similarity with that of a related sea fan species, Eunicella cavolini, with six SNPs and a 49 bp deletion between nad5 and nad4 in E. verrucosa distinguishing the two.
The octocoral Eunicella verrucosa (Pallas, 1766) (common name: pink sea fan) inhabits rocky substrates across the northeast Atlantic and western Mediterranean at depths of 10–200 m; its geographical range stretches from Donegal in northwest Ireland to, reportedly, the coast of Mauritania in West Africa (IUCN, 1996; Hayward & Ryland Citation2017). It has been detrimentally affected by fishing activities across much of its range and is classed as ‘vulnerable’ on the IUCN red list.
The mitochondrial genome was amplified from an individual sea fan collected in September 2008 from East Tennents Reef, Lyme Bay, Dorset, England (N 50° 39.090, W 02° 52.440); the specimen is deposited at the Department of Biosciences, University of Exeter (email: [email protected]) under voucher code Eten04. The genome is one of 17 complete E. verrucosa mitochondrial genomes sequenced in this study; additional specimens originated from sites in the Atlantic, including the west coast of Ireland, southwest Wales, the Isles of Scilly and Dorset (southern England), northwest France, southern Portugal, and the Mediterranean (northeast Spain); see Supplemental Materials Table S1 for details. See Holland et al. (Citation2013, Citation2017) for details of collection protocol and sample preparation.
DNA was extracted using a salting-out protocol modified from that of Li et al. (Citation2011) and Jenkins et al. (Citation2019). phi29-induced rolling circle amplification (RCA) was used to amplify the complete mitochondrial genome; the amplification protocol was modified from that of Dean et al. (Citation2001) and Simison et al. (Citation2006). A sodium acetate-ethanol precipitation was performed to purify the amplified DNA. The circular mtDNA template was then sequenced. Library preparations were carried out using a Mosquito LV (SPT Labtech) using a Nextera XT DNA library preparation kit (Illumina). Individual DNA templates were barcoded and pooled onto a single lane of paired-end 300 bp Illumina MiSeq v2. Mitochondrial genome assembly was conducted as follows: low-quality reads and adaptors were removed using the program Fastq-mcf v.1.04.636; a de novo SPAdes (Nurk et al. Citation2013) assembly was then carried out. De novo contigs produced by SPAdes were blast searched against the complete Eunicella cavolini mitochondrial genome (KY559408). Contigs with significant hits were then extracted and used as a reference against which additional raw reads were assembled. These often comprised one large contig of 19,267 bp, representing the complete mitochondrial genome of E. verrucosa. Additionally, raw reads of E. verrucosa were mapped directly against the mitochondrial genome of E. cavolini (KY559408) using the SAMtools suite of bioinformatic programs. The two genome assembly approaches (de novo-based and aligned-against-a-reference) produced the same mitochondrial genome sequence for E. verrucosa for all specimens analyzed; see Supplemental Material, section S2, for full details.
The 17 complete E. verrucosa mitochondrial genomes sequenced in this study were identical, being 19,267 bp in length, with the same gene arrangement as reported for other species of Eunicella (Brockman and McFadden Citation2012; Poliseno et al. Citation2017), including 14 protein-coding genes (including the mutS gene), two ribosomal RNA subunits (12S and 16S) and one methionine tRNA gene (tRNAmet). Among the protein-coding genes, ten were encoded on the heavy strand (cox1, nad1, coxb, nad6, nad3, nad4L, mutS, nad2, nad5 and nad4) and four were encoded on the light strand (cox2, atp8, atp6 and cox3). Two genes, nad2 and nad5, overlapped by 13 bp; all other genes were separated by intergenic regions (IGRs). All protein-coding genes had the same start codon (ATG) and a TAA or TAG stop codon, except for cox1 which terminated with the incomplete stop codon T--.
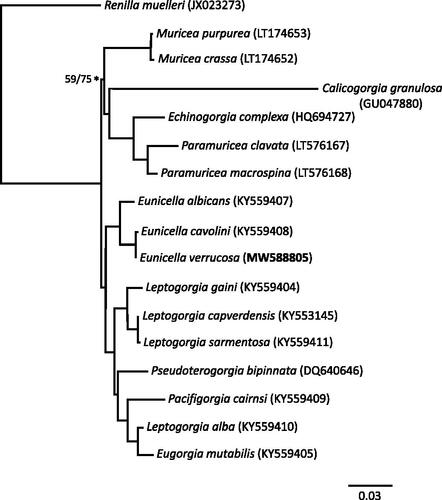
The mitogenome of E. verrucosa showed 99.72% similarity with that of a related sea fan species, Eunicella cavolini, with six SNPs and a 49 bp deletion between nad5 and nad4 in E. verrucosa (compared to E. cavolini) distinguishing the two. Of the six polymorphisms detected between E. verrucosa and E. cavolini, five were in genes (nad6, nad3, nad2, nad4, cox2) and were synonymous; the sixth SNP was in the nad4–cox3 IGR. Even compared to the known low levels of variation observed between the mitogenomes of octocorals (see Poliseno et al. Citation2017 for detailed discussion), the level of variation observed between the mitogenomes of E. verrucosa and E. cavolini is markedly low (<0.3%), though other –morphological and genetic (Holland et al. Citation2013)– data support their designation as separate species. Phylogenetic analysis of E. verrucosa and complete protein-coding sequences from the mitochondrial genomes of other soft coral (subclass Octocorallia) taxa (; Supplemental Material, section S3) placed E. verrucosa with E. cavolini in this phylogeny, though, with complete mitogenome data available for only three out of more than 40 reported Eunicella species (WoRMS Citation2021), exact relationships between them remain to be determined.
Figure 1. Phylogram constructed by maximum-likelihood analysis of 17 octocoral mitochondrial genomes using the PhyML plugin for Geneious v6.1.8. The analysis comprises 16 Alcyonacea (soft corals) genomes and a Pennatulacea (sea pens) genome as an outgroup; sequence data for all 14 mitochondrial protein-coding genes were extracted from complete mitochondrial genomes available in GenBank and aligned using ClustalW in Geneious v6.1.8; see Supplemental Material, section S3, for full details. All nodes (except one – see values for ML/BI on * node) had consensus support values of >70 for maximum likelihood and >90 for Bayesian inference. The Eunicella verrucosa specimen (sequenced in the current study) is labeled in bold.
Supplemental Material
Download MS Word (57.6 KB)Acknowledgements
We are grateful to the following for assistance with sample collection: Jasmin Blenkins-O’Callaghan (Kilkee Dive Centre), Phil Newman (Countryside Council for Wales), Keith Hiscock (Marine Biological Association UK), Julian Turner (University of Exeter Sub-Aqua Club), Laurent Lévêque and staff (Station Biologique de Roscoff), Rita Castilho and staff (University of Algarve – supported by EU ASSEMBLE programme, agreement no.227799) and Andrea Gori (Institut de Ciències del Mar, Barcelona). UK samples were collected under Natural England licences 20080861 and 20090943, and Marine Management licence MMO-0001.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data for this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/nuccore/MW588805 under accession number MW588805.
Additional information
Funding
References
- Brockman SA, McFadden CS. 2012. The mitochondrial genome of Paraminabea aldersladei (Cnidaria: Anthozoa: Octocorallia) supports intramolecular recombination as the primary mechanism of gene rearrangement in octocoral mitochondrial genomes. Genome Biol Evol. 4(9):994–1006.
- Dean FB, Nelson JR, Giesler TL, Lasken RS. 2001. Rapid amplification of plasmid and phage DNA using phi29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 11(6):1095–1099.
- Hayward PJ, Ryland JS. 2017. Handbook of the marine fauna of north-west Europe. 2nd ed. Oxford (UK): Oxford University Press.
- Holland LP, Dawson DA, Horsburgh GJ, Krupa AP, Stevens JR. 2013. Isolation and characterization of fourteen microsatellite loci from the endangered octocoral Eunicella verrucosa (Pallas, 1766). Conservation Genet Resour. 5(3):825–829.
- Holland LP, Jenkins TL, Stevens JR. 2017. Contrasting patterns of population structure and gene flow facilitate exploration of connectivity in two widely distributed temperate octocorals. Heredity. 119(1):35–48.
- Jenkins TL, Ellis CD, Stevens JR. 2019. SNP discovery in European lobster (Homarus gammarus) using RAD sequencing. Conservation Genet Resour. 11(3):253–257..
- Li Y, Wang W, Liu X, Luo W, Zhang J, Gul Y. 2011. DNA extraction from crayfish exoskeleton. Indian J Exp Biol. 49(12):953–957.
- Nurk S, et al. 2013. Assembling genomes and mini-metagenomes from highly chimeric reads. In: Deng M, Jiang R, Sun F, Zhang X, editors. Research in computational molecular biology. RECOMB 2013. Lecture Notes in Computer Science, vol. 7821. Berlin, Heidelberg: Springer..
- Poliseno A, Feregrino C, Sartoretto S, Aurelle D, Wörheide G, McFadden CS, Vargas S. 2017. Comparative mitogenomics, phylogeny and evolutionary history of Leptogorgia (Gorgoniidae). Mol Phylogenet Evol. 115:181–189.
- Simison WB, Lindberg DR, Boore JL. 2006. Rolling circle amplification of metazoan mitochondrial genomes. Mol Phylogenet Evol. 39(2):562–567.
- WoRMS. 2021. World Register of Marine Species - Eunicella Verrill, 1869. marinespecies.org. Retrieved 17/09 2021.
