Abstract
Dictyosquilla foveolata is a moderately important commercial species that is naturally distributed from Australia to Borneo, and from northwest Indonesia to Burma, Vietnam, and China. The complete mitogenome sequence of D. foveolata was first determined using Illumina HiSeq sequencing in this study. The circular genome was 15,733 bp in length and consisted of 13 protein-coding genes and 2 ribosomal RNA genes, 22 transfer RNA genes. The mitochondrial gene arrangement of D. foveolata is similar to those of most other stomatopods. Results from maximum-likelihood phylogenetic analysis showed that D. foveolata was clustered with other six species of the family Squillidae. This study will be valuable for phylogenetic analyses of the genus Dictyosquilla and the other genera of the order Stomatopoda. This is the first record of the complete mitogenome for the genus Dictyosquilla.
Dictyosquilla foveolata (Wood-Mason, 1895) is a moderately important commercial species that is naturally distributed from Australia to Borneo, and from northwest Indonesia to Burma, Vietnam, and China, and always inhabits sandy-muddy bottoms in water less than 50 m in depth (Huang et al. Citation2009). As the life history of the order Stomatopoda includes multiple pelagic larval stages, more molecular markers than the typical COXI are necessary for identifying them to species (Feller et al. Citation2013). However, the taxonomic group comprises over 480 named species (Kundu et al. Citation2018), but only a few complete mitochondrial genomes of Stomatopoda have been determined up to the present (Zhang et al. Citation2020). The D. foveolata mitogenome here provided could be used both as a tool for larval identification as well as a source of information for phylogenetic analyses.
The samples of D. foveolata were collected from the offshore sea of Ganjiang Town in Yuhuan County, Taizhou City, Zhejiang Province (28°01′ N, 121°40′ E). Total genomic DNA of D. foveolata was extracted from the muscle tissue using E.Z.N.A®DNA kit (OMEGA, USA), and a specimen was deposited at Specimen Museum of Marine Fisheries Research Institute of Zhejiang (Shanshan Zhou and [email protected]) under the voucher number ZMFRI20210301. Mitochondrial DNA was amplified with a DNA REPLI-g Mitochondrial DNA Kit (QIAGEN, Hilden, Germany) as directed by the manufacturer. The library construction and sequencing were performed by Biozeron (Biozeron, Shanghai, China) using the Illumina HiSeq 4000 sequencing platform (Illumina, San Diego, CA). The software SPAdes (3.10.1) (Bankevich et al. Citation2012) was employed to assemble the mitogenome, and the sequences from two different individuals were assembled independently for confirming the accuracy of the results.
The complete mitochondrial genome sequence of D. foveolata has been deposited in GenBank with accession no. MW864094. The circular genome (15,733 bp) comprised 13 protein-coding genes (PCGs), 2 rRNA genes (12S rRNA and 16S rRNA), 22 transfer RNA (tRNA) genes. The nucleotide composition of the heavy strand of D. foveolata was 36.65% for A, 17.75% for C, 12.57% for G, and 33.03% for T, total AT content was up to 69.68%. On one hand, six PCGs (COXII, COXIII, CYTB, ATP6, ND4 and ND4L) were initiated with ATG codon, and the remaining PCGs were initiated with ATT (ND2), GTG (ND3), ATC (COXI, ATP8) or ATA (ND1, ND5 and ND6). On the other hand, all the PCGs were terminated with TAA codon, except for COXI, COXII and ND6, terminated with T. The two ribosomal RNA genes, 12S rRNA gene (826 bp) and 16S rRNA gene (1356 bp), were encoded on the light strand. Besides, eight tRNAs (tRNATyr, tRNAGln, tRNAVal, tRNALeu, tRNAPro, tRNAHis, tRNAPhe and tRNAcys) and four PCGs (ND1, ND4, ND4L and ND5) were located on the light strand, whereas the other tRNAs and PCGs were all located on the heavy strand.
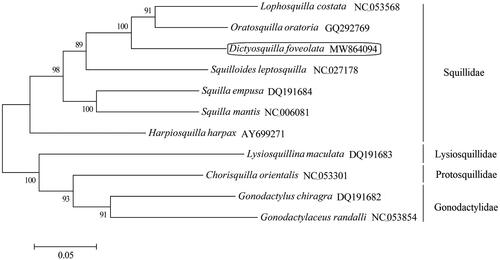
A total of 11 complete mitogenome sequences from the same order Stomatopoda aligned by CLUSTRALW have been used to construct a phylogenetic tree () by the maximum-likelihood method based on the Tamura-Nei model (MEGA7 software) (Kumar et al. Citation2016). In the phylogenetic tree, all the stomatopods were separated into two branches, one comprising the family Squillidae and the other comprising the family Lysiosquillidae, Protosquillidae and Gonodactylidae. D. foveolata was clustered into one clade with other six species from Squillidae. The phylogenetic tree also provided a resource for estimating the evolutionary history of the order Stomatopoda.
Disclosure statement
The authors report no conflicts of interest. The authors are responsible for the content and writing of the paper.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] under the accession no. MW864094. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA736567, SRR14777097, and SAMN19653974 respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Feller KD, Cronin TW, Ahyong ST, Porter ML. 2013. Morphological and molecular description of the late-stage larvae of Alima Leach, 1817 (Crustacea: Stomatopoda) from Lizard Island, Australia. Zootaxa. 3722(1):22–32.
- Huang J, Yang T, Wang B. 2009. Spatio-temporal patterns of stomatopods (Malacostraca, Stomatopoda) in the main bays of Guangdong Province, China. Crustac. 82(8):1029–1043.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Kundu S, Rath S, Tyagi K, Chakraborty R, Pakrashi A, Kumar V, Chandra K. 2018. DNA barcoding of Cloridopsis immaculata: genetic distance and phylogeny of stomatopods. Mitochondrial DNA B Resour. 3(2):955–958.
- Zhang Y, Bi Y, Feng M. 2020. The complete mitochondrial genome of Lophosquillia costata (Malacostraca: Stomatopoda) from China and phylogeny of stomatopods. Mitochondrial DNA B Resour. 5(3):2495–2497.