Abstract
The complete cp genome of Prunus tangutica is 158,131 bp in length, exhibits a typical quadripartite structural organization, consisting of a large single copy (LSC) region of 86,266 bp, two inverted repeats (IR) regions of 26,389 bp, and a small single copy (SSC) region of 19,087 bp. The cp genome contains 131 complete genes, including 86 protein-coding genes (86 PCGs), 8 ribosomal RNA genes (8 rRNAs), and 37 tRNA genes (37 tRNAs). Most genes occur in a single copy, while 19 genes occur in double, including 4 rRNAs (4.5S, 5S, 16S, and 23S rRNA), 7 tRNAs (trnA-UGC, trnI-GAU, trnL-CAA, trnI-CAU, trnN-GUU, trnR-ACG, and trnV-GAC), and 5 PCGs (rps7, ndhB, ycf2, rpl2 and rpl23). The overall GC content of cp DNA is 36.7%, the corresponding values of the LSC, SSC, and IR regions are 34.6%, 30.1%, and 42.6%, respectively. Further, the phylogenetic analysis suggested that the P. tangutica was closely related to Prunus tenella. The results of P. tangutica will lay a foundation for further research.
Introduction
Prunus L. belongs to the subfamily Amygdaloideae of the Rosaceae. It consists of 200 species with most species in the temperate zone (Yu et al. Citation1986; Ghora and Panigrahi Citation1995; Mabberley Citation1997). Prunus tangutica (Batal.) Korsh. is economically important because many species are sources of oil, timber, and ornamentals (Sangtae and Jun Citation2001). However, due to anthropogenic overexploitation and decreasing distributions, this species needs urgent conservation. Knowledge of the genetic information about this species would contribute to the formulation of a protection strategy. In this study, we assembled the complete chloroplast genome of P. tangutica, hoping to lay a foundation for further research.
Fresh leaves of P. tangutica were collected from the psammophyte germplasm bank of Yinchuan Botanical Garden (Yinchuan, Ningxia, China; coordinates: 105°49′18″E, 38°08′42″N) and dried with silica gel. The voucher specimen was stored in the State Key Laboratory of Seeding Bioengineering with the number is NFILSBZJ20210116. Plant Chloroplast Genomic DNA was extracted with a modified CTAB method (Doyle and Doyle Citation1987). We used Illumina HiSeq X Ten sequencing and MITObim v1.9 program of Hahn et al. (Citation2013), with the help of close reference sequences to assemble chloroplast genomes. We assembled the complete chloroplast genome by GetOrganelle pipeline v1.6.3a (Jin et al. Citation2020). Plann v1.1 (Huang and Cronk Citation2015), Geneious v11.0.3 (Kearse et al. Citation2012) were used to annotate the chloroplasts genome and correct the annotation. The OGDraw online tool (Lohse et al. Citation2013) (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html) was used to produce a genome map. The complete cp genome was deposited in GenBank (accession number: MZ145044)
The total plastome length of P. tangutica is 158,131 bp, exhibits a typical quadripartite structural organization, consisting of a large single copy (LSC) region of 86,266 bp, two inverted repeats (IR) regions of 26,389 bp, and a small single copy (SSC) region of 19,087 bp. The cp genome contains 131 complete genes, including 86 protein-coding genes (86 PCGs), 8 ribosomal RNA genes (8 rRNAs), and 37 tRNA genes (37 tRNAs). Most genes occur in a single copy, while 19 genes occur in double, including 4 rRNAs (4.5S, 5S, 16S, and 23S rRNA), 7 tRNAs (trnA-UGC, trnI-GAU, trnL-CAA, trnI-CAU, trnN-GUU, trnR-ACG, and trnV-GAC), and 5 PCGs (rps7, ndhB, ycf2, rpl2 and rpl23). The overall GC content of cp DNA is 36.7%, the corresponding values of the LSC, SSC, and IR regions are 34.6%, 30.1%, and 42.6%.
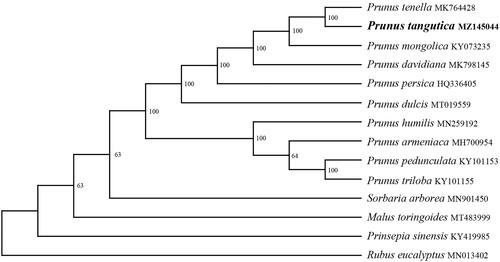
In order to further clarify the phylogenetic position of P. tangutica, a plastome of 13 representatives Prunus species were obtained from NCBI to reconstruct the plastome phylogeny, with Sorbaria arborea, Malus toringoides, Prinsepia sinensis, and Rubus eucalyptus as an outgroup. All the sequences were aligned using MAFFT v.7.313 (Katoh and Standley Citation2013) and maximum likelihood phylogenetic analyses were conducted using RAxML v.8.2.11 under GTRCAT model with 500 bootstrap replicates. The phylogenetic tree shows that the species of Prunus were divided into two subclades (). P. humilis, P. armeniaca, P. pedunculata, and P. triloba clustered together, and remain species clustered in another clade. while P. tangutica is a sister to P. tenella.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that obtained at this study are openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov/) under accession number of MZ145044. The associated BioProject and SRA numbers are PRJNA762480 and SUB10364517, respectively.
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Ghora C, Panigrahi G. 1995. The family Rosaceae in India. Vol. 2. Dehra Dun: Bishen Singh Mahendra Pal Singh.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucl Acids Res. 41(13):e129–e129.
- Huang D, Cronk Q. 2015. Plann: a command-line application for anno-tating plastome sequences. Appl Plant Sci. 3(8):1500026.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Katoh K, Standley D. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W81.
- Mabberley D. 1997. The plant-book, a portable dictionary of the vascular plants. 2nd ed. Cambridge (UK): Cambridge University Press.
- Sangtae L, Jun W. 2001. A phylogenetic analysis of Prunus and the Amygdaloideae (Rosaceae) using ITS sequences of nuclear ribosomal DNA. Am J Bot. 88:150–160.
- Yu T, Lu L, Ku T, Li C, Chen S. 1986. Rosaceae (3), amygdaloideae. In: Flora reipublicae popularis sinicae. Vol. 38. Beijing (China): Science Press.