Abstract
Rusa unicolor has been listed as a vulnerable species by the International Union for Conservation of Nature and Natural Resources because of human activities. In recent years, population numbers have decreased due to heavy hunting and habitat loss, and little genetic data on this species exists; thus, our knowledge of range distribution and population size remains limited. In the current study, the complete R. u. cambojensis mitochondrial genome was sequenced using polymerase chain reaction followed by direct sequencing. The complete mitochondrial genome was determined to be circular and contain 16,557 bp, including 13 protein-coding genes, 2 rRNA genes, 22 tRNA genes, and 1 control region, the gene composition and order were similar to those of most other vertebrates reported to date. Most mitochondrial genes, except for ND6 and eight tRNAs, were encoded on the heavy strand. The overall base composition of the heavy strand was 33.6% A, 28.9% T, 24.2% C, and 13.3% G, with a strong AT bias of 62.5%. There were 13 regions of gene overlap totaling 96 bp and 12 intergenic spacer regions totaling 70 bp. The phylogenetic analyses (maximum likelihood and Bayesian inference) of R. unicolor based on the mitochondrial genome four subspecies of R. unicolor were clustered into a well-supported single clade, and R. u. cambojensis was most closely related to R. u. dejeani. This study will assist in the exploration of the evolutionary history and taxonomic status of the sambar, as well as its protection as a genetic resource.
Rusa unicolor (Kerr 1792, Artiodactyla: Cervidae) or the sambar is the most widespread deer in Asia (Pocock Citation1943; Grubb Citation1990; Leslie Citation2011). It is listed as vulnerable on the International Union for Conservation of Nature and Natural Resources Red List because of its reduced range (Timmins et al. Citation2015). Characteristics of single genes (e.g. mitochondrial 16S rRNA, Cytb, and COI) have been used to distinguish R. unicolor from other species (Chen et al. Citation1991; Liu et al. Citation2003; Guha and Kashyap Citation2005; El-Jaafari et al. Citation2008; Kumar et al. Citation2012; Cai et al. Citation2016). The mitochondrial genome is better for investigating species evolution and population genetics (Hassanin et al. Citation2012). Within the seven subspecies, only the genomes of R. u. swinhoei, R. u. dejeani, and R. u. hainana are available (Chen et al. Citation2011; Wu et al. Citation2016; Liu et al. Citation2019). In the current investigation, the complete mitochondrial genome of R. u. cambojensis was sequenced and characterized to provide fundamental molecular data for further conservation and phylogenetic studies of this large mammal. Possible relationships between subspecies of R. unicolor are also discussed.
One dead R. u. cambojensis specimen was collected from Naban River Watershed National Nature Reserve (100°42′ E, 22°14′ N), Xishuangbanna, Yunnan Province, southwestern China on 8th July 2017. The sample (1707001) was deposited in the Center for Integrative Conservation, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Mengla, Yunnan province, China. Total genomic DNA was extracted from tissue using a DNeasy Blood & Tissue Kit (Tiangen Biochemistry Technology Co., Ltd., China) and sequenced using an ABI PRISM 3700 sequencing system (Applied Biosystems, Foster City, CA, USA). To obtain the whole mitogenomic sequence, 22 pairs of primers were designed according to previous studies (Chen et al. Citation2011; Hassanin et al. Citation2012; Wu et al. Citation2016; Liu et al. Citation2019). The base composition of the mitochondrial genome was analyzed using MEGA 5.05 (Tamura et al. Citation2011). The genome sequence was annotated using DOGMA (Wyman et al. Citation2004) and was deposited on the NCBI website (https://www.ncbi.nlm.nih.gov/genbank/) with the accession number MK941883. Features of the complete mitochondrial genome of R. u. cambojensis were identical to those of other sambar subspecies (e.g. Chen et al. Citation2011; Wu et al. Citation2016; Liu et al. Citation2019). It was a circular molecule, 16,557 bp in length, and included 13 protein-coding genes, 2 rRNA genes, 22 tRNA genes, and 1 non-coding control region (D-loop). The overall base composition of the heavy strand was 33.6% A, 28.9% T, 24.2% C, and 13.3% G, with a strong AT bias of 62.5%.
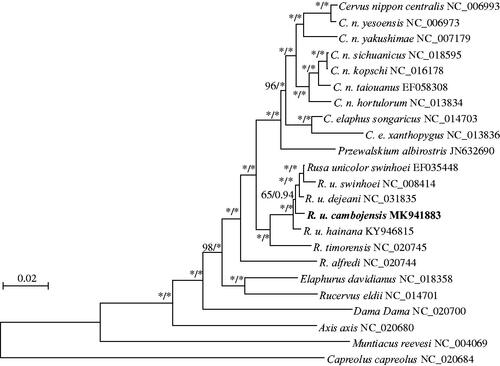
Phylogenies of the mitogenome were constructed using maximum likelihood, implemented in PHYML 3.0 (Guindon et al. Citation2010). Bayesian inference was implemented in MRBAYES 3.2.1 (Ronquist et al. Citation2012). Based on the complete genomes (), four subspecies of R. unicolor were clustered in a well-supported single clade, with R. timorensis as its sister species. Intraspecific phylogenetics demonstrated that R. u. cambojensis was more closely related to R. u. dejeani than to R. u. swinhoei and R. u. hainana, which are respectively endemic to the islands of Taiwan and Hainan. The R. u. cambojensis mitogenome will be useful for its identification and conservation, as well as for evolutionary research on R. unicolor.
Figure 1. Maximum likelihood and Bayesian inference phylogenetic trees for Rusa unicolor based on 23 complete mitochondrial genomes. Numbers on branches indicate bootstrap support for maximum likelihood, followed by the posterior probability in Bayesian inference analyses for the node. Stars indicate values of 100 for maximum likelihood and 1.00 for Bayesian inference.
Acknowledgments
Laboratory work was conducted at the Central Laboratory, Public Technology Service Center, Xishuangbanna Tropical Botanical Garden (XTBG), Chinese Academy of Sciences (CAS). The authors sincerely thank Ms. Nan Sun for proofreading the manuscript.
Disclosure statement
The authors declare no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The datasets supporting the results of this article are available in GenBank of the NCBI at (https://www.ncbi.nlm.nih.gov/) under accession number MK941883.
Additional information
Funding
References
- Cai YS, Zhang L, Wang YW, Liu Q, Shui QL, Yue BS, Zhang ZH, Li J. 2016. Identification of deer species (Cervidae, Cetartiodactyla) in China using mitochondrial cytochrome c oxidase subunit I (mtDNA COI)). Mitochondrial DNA A DNA Mapp Seq Anal. 27(6):4240–4243.
- Chen CH, Huang HL, Chang MT, Chiang LC, Cheng SL, Liu BT, Wang CH, Wu MC, Cronin MA, Palmisciano DA, Vyse ER, et al. 1991. Mitochondrial DNA in wildlife forensic science: species identification of tissues. Wildl Soc Bull. 19:94–105.
- Chen C-H, Huang H-L, Chang M-T, Chiang L-C, Cheng S-L, Liu B-T, Wang C-H, Wu M-C, Huang M-C. 2011. Characterization of mitochondrial genome of Formosan sambar (Rusa unicolor swinhoei). Biologia. 66(6):1196–1201.
- El-Jaafari HAA, Panandam JM, Idris I, Siraj SS. 2008. RAPD analysis of three deer species in Malaysia. Asian Australas J Anim Sci. 21(9):1233–1237.
- Grubb P. 1990. List of deer species and subspecies. Deer, Journal of the British Deer Society. 8:153–155.
- Guha S, Kashyap VK. 2005. Development of novel heminested PCR assays based on mitochondrial 16S rRNA gene for identification of seven pecora species. BMC Genet. 6:42–49.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321.
- Hassanin A, Delsuc F, Ropiquet A, Hammer C, Jansen van Vuuren B, Matthee C, Ruiz-Garcia M, Catzeflis F, Areskoug V, Nguyen TT, et al. 2012. Pattern and timing of diversification of Cetartiodactyla (Mammalia, Laurasiatheria), as revealed by a comprehensive analysis of mitochondrial genomes. C R Biol. 335(1):32–50.
- Kumar US, Ratheesh RV, Thomas G, George S. 2012. Use of DNA barcoding in wildlife forensics: a study of sambar deer (Rusa unicolor). Forest Science and Technology. 8(4):224–226.
- Leslie DM. 2011. Rusa unicolor (Artiodactyla: Cervidae). Mammalian Species. 43(1):1–30.
- Liu HT, Dong YM, Xing XM, Yang FH. 2019. Characterization of the complete mitochondrial genome of Rusa unicolor hainana (Artiodactyla: Cervidae). Conservation Genet Resour. 11 (2):143–144.
- Liu XY, Wang YQ, Liu ZQ, Zhou KY. 2003. Phylogenetic relationship of Cervinae based on sequence of mitochondrial cytochrome b gene. Zoological Research. 24(1):27–33.
- Pocock RI. 1943. The larger deer of British India. Part III-the sambar (Rusa). Journal of the Bombay Natural History Society. 44:27–37.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28(10):2731–2739.
- Timmins RKK, Giman B, Lynam A, Chan B, Steinmetz R, Sagar BH, Samba KN. 2015. Rusa unicolor. IUCN Red list of threatened species (Version 2017–1). International Union for Conservation of Nature and Natural Resources. http://www.iucnredlist.org.
- Wu XY, Qi Y, Li B, Yao YF, Xu HL. 2016. The complete mitochondrial genome sequence of Rusa unicolor dejeani (Artiodactyla: Cervidae). Conservation Genet Resour. 8(3):255–257.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
