Abstract
Cinnamomum tenuipile Kosterm is a precious aromatic tree in Lauraceae. To better determine its phylogenetic location with other Cinnamomum species, the chloroplast genome of C. tenuipile was sequenced. The complete chloroplast genome size is 152,761 bp, consisting of a pair of inverted repeats (IRa/b) with a length of 20,074 bp separated by a large single-copy region (LSC) and a small single-copy region (SSC) which are 93,685 and 18,928 bp, respectively. The overall GC content of the cp genome is 39.16%. The maximum-likelihood phylogenetic tree showed that C. tenuipile is more closely related to C. aromaticum, providing new insight into the evolution of Lauraceae.
Cinnamomum tenuipile is an aromatic tree species in Lauraceae that native to southwestern China. The essential oils distilled from C. tenuipile contain 86–98% geraniol, which has economic importance in the spice industry (Yang et al. 2005). However, excessive logging leads to the reduction of natural resources of C. tenuipile. In order to better protect and understand the phylogeny of C. tenuipile, the chloroplast genome has been studied based on high-throughput sequencing approaches.
DNA was isolated from fresh young leaves of C. tenuipile at Xishuangbanna (21.69°N,100.06°E). The voucher specimen (ZSS-ZYJ-20200830) was deposited at Camphor Engineering Technology Research Center for National Forestry and Grassland Administration (Yongjie Zheng, [email protected]). A 350 bp library was constructed and sequenced by using the Illumina NovaSeq platform. Raw reads were polished and assembled by SPAdes (version: 3.13.0) (Bankevich et al. Citation2012) with the reference genome sequence of Cinnamomum camphora (GenBank: NC_035882) (Chen et al. 2017). The complete chloroplast (cp) genome sequence has been published on the GenBank with accession number: MW421304.
The cp genome size of C. tenuipile was 152,761 bp, including an LSC region of 93,685 bp, an SSC region of 18,928 bp, and a pair of IR regions of 20,074 bp. The overall GC content is 39.14% (LSC, 37.96%; SSC, 33.91%; IR, 44.44%). The cp genomes were annotated with 131 genes, including 83 protein-coding genes, 44 tRNA genes, and 4 rRNA genes.
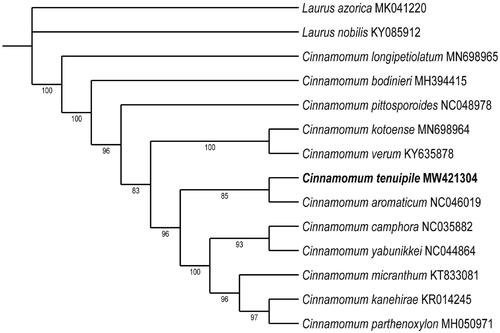
A phylogenetic tree was reconstructed to confirm the phylogenetic location of C. tenuipile, with Laurus species as an outgroup (). The complete chloroplast genome sequences were aligned by using MAFFT (Katoh et al. 2002). Maximum likelihood (ML) phylogenetic analyses were performed based on K3Pu + F + I + G4 model by ModelFinder and iqtree (version:1.6.7) with 1000 bootstrap replicates (Nguyen et al. 2015; Kalyaanamoorthy et al. 2017; Hoang et al. 2018). The ML phylogenetic tree with 83%–100% bootstrap values supported that C. tenuipile and C. aromaticum were in the same clade. The complete chloroplast genome of C. tenuipile provides valuable genomic resources for improving our understanding of species phylogeny in Lauraceae, exploring genetic variations and designing conservation strategies.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/) under the accession no. MW421304. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA749606, SAMN20394009, and SRR15243023, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Chen C, Zheng Y, Liu S, Zhong Y, Wu Y, Li J, Xu L-A, Xu M. 2017. The complete chloroplast genome of Cinnamomum camphora and its comparison with related Lauraceae species. PeerJ. 5:e3820.
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Yang T, Li J, Wang HX, Zeng Y. 2005. A geraniol-synthase gene from Cinnamomum tenuipilum. Phytochemistry. 66(3):285–293.