Abstract
Callicarpa longifolia Lamk. var. floccosa Schauer is a species with medicinal and ornamental values. In this study, the complete chloroplast genome of C. longifolia var. floccose is reported. The chloroplast genome of this species is 154,285 bp in length and contains a typical circular quadripartite structure. There are two inverted repeats of 25,700 bp, which is separated by a large single-copy region of 85,008 bp and a small single-copy region of 17,877 bp. The complete chloroplast contains 112 distinct genes, including 78 protein-coding, 30 tRNAs and 4 rRNAs genes. Phylogenetic analysis suggests that C. longifolia var. floccose is closely associated with C. formosana.
Callicarpa longifolia Lamk. var. floccosa Schauer 1847 is a perennial shrub of genus Callicarpa and family Lamiaceae (Chen and Michael Citation1994, Li et al. Citation2016). It is mainly distributed in southwestern China, India, Singapore, Indonesia, and the Philippines. This species has high ornamental value with clustered white, purple or red berries, and also has certain medicinal value. In this study, the chloroplast (cp) genome characteristics of C. longifolia var. floccose were assembled and described for the first time. It will provide valuable resources for further study of genetic conservation, evolution, and molecular breeding studies in the genus of Callicarpa L.
Fresh leaves of C. longifolia var. floccose was collected from Pengshui County of Chongqing, China (N29°00′47.85″, E107°52′31.09″, 553 m). The voucher specimen (CY Wang PS 20191002) was deposited in the herbarium of Southwest University (SWCTU, Qian Wang, [email protected]). Whole cp genome sequences were generated by using Illumina HiSeq 4000 platform at the Beijing Novogene Bioinformatics Technology Co., Ltd. (Nanjing, China). To obtain the complete cp genome, de novo assembly of the raw data was performed by using SPAdes version 3.5.0 (Bankevich et al. Citation2012). The cp genome sequence was annotated by using PGA (https://github.com/quxiaojian/PGA) (Qu et al. Citation2019) with manual adjustments. The sequence cp genome was deposited in GenBank (accession number MW149076).
The cp genome of C. longifolia var. floccose formed a typical circular quadripartite structure of 154,285 bp length, containing two inverted repeats (IRs) of 25,700 bp, a large single-copy (LSC) region of 85,008 bp, and a small single-copy (SSC) region of 17,877 bp. The GC content of the whole cp genome is 38.07%. In total, 112 distinct genes were annotated, including 78 protein-coding (PCGs), 30 transfer RNA (tRNA), and 4 ribosomal RNA (rRNA) genes. The gene content and organization are similar to other members of Callicarpa (Wang et al. Citation2019; Du et al. Citation2020; Wang et al. Citation2021; Xie et al. Citation2021).
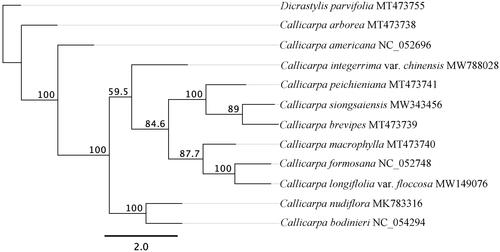
To determine the phylogenetic position of C. longifolia var. floccose with related species, a phylogenetic analysis was performed based on 11 complete cp genomes of Callicarpa. In addition, Dicrastylis parvifolia (MT473755) was included as an outgroup. The sequences were aligned by MAFFT version 7.0 (Katoh and Standley Citation2013). The maximum-likelihood (ML) phylogenetic trees were reconstructed by using RAxML version 8.2.11 (Stamatakis Citation2014) with the GTR Gamma and 1,000 bootstrap replicates. As shown in the phylogenetic tree (), C. longifolia var. floccose was most closely to C. formosana.
Figure 1. A maximum likelihood (ML) tree of genus Callicarpa based on twelve complete chloroplast genome sequences. Numbers at nodes correspond to ML bootstrap percentages (1,000 replicates). All the sequences are available in GenBank, with the accession numbers listed right to their scientific names.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Chloroplast data supporting this study are openly available in GenBank at nucleotide database, https://www.ncbi.nlm.nih.gov/nuccore/MW149076, Associated BioProject, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA757227, BioSample accession number at https://www.ncbi.nlm.nih.gov/biosample/SAMN20955945 and Sequence Read Archive at https://www.ncbi.nlm.nih.gov/sra/SRR15584543.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Chen SL, Michael GG. 1994. Flora of China: Verbenaceae. Vol.17. Beijing: Science Press; St. Louis: Missouri Botanical Garden Press; p. 1–49.
- Du YX, Liu YF, Liu B, Wang TL. 2020. Complete chloroplast genome of Callicarpa formosana Rolfe, a famous ornamental plant and traditional medicinal herb. Mitochondr DNA B Resour. 5(3):3383–3384.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Li B, Cantino PD, Olmstead RG, Bramley GLC, Xiang CL, Ma ZH, Tan YH, Zhang DX. 2016. A large-scale chloroplast phylogeny of the Lamiaceae sheds new light on its subfamilial classification. Sci Rep. 6(1):34343.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):1–12.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wang CY, Luo JH, He HL, Wang Q. 2021. The complete chloroplast genome of Callicarpa bodinieri (Lamiales, Lamiaceae), an ornamental and medicinal plant from Chongqing, China. Mitochondr DNA B Resour. 6(3):1229–1230.
- Wang HX, Chen L, Cheng XL, Chen WS, Li LM. 2019. Complete plastome sequence of Callicarpa nudiflora Vahl (Verbenaceae): a medicinal plant. Mitochondr DNA B Resour. 4(2):2090–2091.
- Xie YQ, Luo ST, Zhang LT, Huang H, Zhang Q, Lai MY, Deng CY. 2021. The complete chloroplast genome of Callicarpa siongsaiensis Metcalf (Lamiaceae) from Fujian Province, China: genome structure and phylogenetic analysis. Mitochondrial DNA B Resour. 6(5):1634–1635.
