Abstract
Adenophora kayasanensis Kitam., belonging to the family Campanulaceae, is an important species because it is used as a type of herbal medicine and is endemic to Korea. Here, we report the complete chloroplast genome sequence of A. kayasanensis as determine by means of Illumina high-throughput sequencing. The complete cp genome was 169,433 bp in length, containing a large single copy (LSC) of 123,110 bp and a small single copy (SSC) of 8619 bp, which were separated by a pair of 29,085 bp inverted repeats (IRs). A total of 112 unique genes were annotated, consisting of 78 protein-coding genes, 30 tRNA genes, and four rRNA genes. The overall GC content is 37.7%. A maximum-likelihood (ML) tree based on 76 protein-coding genes indicated that A. kayasanensis is closely related to Adenophora racemosa. This newly sequenced chloroplast genome will be useful to those engaged in research on the phylogenetic position of A. kayasanensis and the evolution of the genus Adenophora.
The genus Adenophora, commonly called ‘Radix Adenophora’, is an important plant resource as a source of herbal medicines (Ji et al. Citation2010). This genus, which belongs to Campanulaceae, is a perennial herbaceous plant genus with approximately 50–100 species that are distributed in temperate regions in Eurasia (Cheon et al. Citation2017). Among Adenophora species, Adenophora kayasanensis Kitam., discussed in this study, is a species endemic to Korea and was first described by Kitamura (Citation1936) after its collection from Mt. Kaya National Park in Korea. In previous phylogenetic studies (Kim and Yoo Citation2011, Citation2012), the phylogenetic relationships of A. kayasanensis were not clear because it exhibited unresolved paraphyly with related taxa. In this study, we report the complete chloroplast genome sequence of A. kayasanensis for the first time, providing useful genetic information for further studies of the phylogenetic position of A. kayasanensis and the evolution of the genus Adenophora.
Plant materials of A. kayasanensis were collected from Mt. Kaya (37°28′17′′N, 130°54′03.77′′E), Hapcheon-gun, Gyeongsangnam-do province in South Korea, and a voucher specimen and DNA sample were deposited as Kangwon National University Herbarium (voucher no. KWNU93474; KS Cheon, [email protected]). The total genomic DNA was extracted from A. kayasanensis leaves using a DNeasy Plant Mini Kit (Qiagen Inc., Valencia, CA) and paired-end (PE) libraries were prepared using a TruSeq DNA Sample Prep Kit HT (Illumina Inc, San Diego, CA) with a 350 bp average insert size. PE libraries were sequenced on a MiSeq platform (Illumina). We obtained 8,205,714 raw reads with a length of 301 bp. Low-quality sequences (Phred score < 20) were trimmed using CLC Genomics WorkBench version 6.04 (CLC Inc., Arhus, Denmark). After trimming, the library for A. kayasanensis included 6,539,849 reads. Then, de novo assembly was implemented using the Geneious prime®2021.1.1 (Biomatters Ltd, Auckland, New Zealand) and a total of 170,095 reads were aligned. We also compared each gene to the published complete chloroplast genome sequence of Adenophora species for correct gene annotation.
A typical quadripartite structure of A. kayasanensis is a DNA molecule 169,433 bp in length with 37.7 GC content, composed of a large single copy (LSC) region of 123,110 bp, a small single copy (SSC) region of 29,085 bp, and two inverted-repeat (IR) regions of 8619 bp. The plastid genome contains a total of 112 unique genes consisting of 78 protein-coding genes, 30 transfer RNA (tRNA) genes, and four ribosomal RNA (rRNA) genes. Of which, two genes contain two introns, and 15 genes contain one intron.
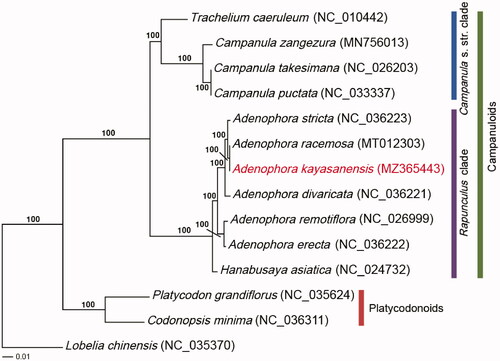
Phylogenetic analyses were conducted with 76 protein-coding genes of 14 accessions. Thirteen Campanulaceae s. str. species were selected as the ingroups, and one species (Lobelia chinensis) was chosen as the outgroup. The sequences were aligned using MAFFT (Katoh and Standley Citation2013). A maximum-likelihood (ML) analysis was performed using RAxML version 7.4.2 with 1000 bootstrap replicates and the GTR + I + Γ model (Stamatakis Citation2006). The ML tree formed the following two clades: platycodonoids and campanuloids. The campanuloids formed two subclades: the Campanula s. str. clade and the Rapunculus clade. All nodes in the ML tree were strongly supported, with 100% bootstrap value (BP) (). Furthermore, Genus Adenophora forms a monophyletic and A. kayasanensis has a closer relationship with A. racemosa than with A. divaricata. In the light of the results of this study, therefore, it is judged that the taxonomic identity of A. kayasanensis will become clear if in-depth studies on the taxonomic relationships with A. racemosa are conducted in the future.
Disclosure statement
The authors declare no conflict of interest. The authors alone are responsible for the content and writing of the article.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MZ365443. The associated BioProject, SRA, and bio-sample numbers are PRJNA736621, SRR14777593, and SAMN19655181, respectively.
Additional information
Funding
References
- Cheon KS, Kim KA, Yoo KO. 2017. The complete chloroplast genome sequences of three Adenophora species and comparative analysis with Campanuloid species (Campanulaceae). PLoS One. 12(8):e0183652.
- Ji YJ, Moon BC, Lee AY, Chun JM, Choo BK, Kim HK. 2010. Molecular phylogenetic position of Adenophora racemosa, an endemic species in Korea. Korean J Crop Sci. 18:379–388.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kim KA, Yoo KO. 2011. Phylogenetic relationships of Korean Campanulaceae based on PCR-RFLP and ITS sequences. Korean J Pl Taxon. 41(2):119–129.
- Kim KA, Yoo KO. 2012. Phylogenetic relationships of Korean Campanulaceae based on chloroplast DNA sequences. Korean J Pl Taxon. 42(4):282–293.
- Kitamura S. 1936. Notes on some Adenophora of Japan. Acta Phytotax Geobot. 5:204–210.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690.