Abstract
Hordeum distichon (H. distichon) is a two-row cultivated barley used as food and as a feed crop. Chloroplast genome is an excellent way to study the genetic structure and evolutionary process of natural population of plant species in recent years. In this study, the complete chloroplast genome of H. distichon was sequenced and analyzed: the size of the chloroplast genome is 136,462 bp in length, including a large single copy region (LSC) of 80,597 bp, a small single copy region (SSC) of 12,701 bp, and a pair of inverted repeated regions (IR) of 21,582 bp; the H. distichon chloroplast genome encodes 129 genes, including 83 protein-coding genes, 38 tRNA genes, and eight rRNA genes; the overall GC-content of the chloroplast genome was 38.32%, with the LSC, SSC, and IR regions being 36.31%, 32.33%, and 43.83%, respectively. Phylogenetic analysis based on 32 species with the maximum likelihood (ML) method indicated that H. distichon was closely related to Hordeum vulgare.
Hordeum distichon (H. distichon), commonly known as two-row cultivated barley, belongs to the genus Hordeum of the family Poaceae which is the important cereal crop grown in nearly all the cultivated areas of the world (Carpici and Celik Citation2012). H. distichon is used for malting, and the best malt quality for beer is produced from two-row cultivated varieties (Bizuneh and Abebe Citation2019). Golden Promise is considered as a two-row cultivated barley variety of H. distichon, which is widely used in brewery barley breeding because of its high yield and excellent brewery quality (Schreiber et al. Citation2019; Reichel et al. Citation2021).
The chloroplast genome is smaller than the nuclear genome, and compared with mitochondrial genome, chloroplast genome structure is relatively conservative (Niu et al. Citation2020). Therefore, chloroplast genome is an excellent way to study the genetic structure and evolutionary process of natural population of plant species in recent years. Chloroplast genome has been widely used in plant research (Su et al. Citation2020).
In this study, the complete chloroplast genome of H. distichon was sequenced and analyzed by using the two-row cultivated barley variety Golden Promise as the research material. The fresh leaves of Golden Promise were collected from Qinghai University artificial climate incubator in Xining, Qinghai, China (101°78′ E, 36°62′ N) and the DNA of Golden Promise was deposited at the Qinghai Key Laboratory of Hulless Barley Genetics and Breeding (Xin Li, [email protected]) under the storage number HG-20201230-01. The genomic DNA was extracted following the modified cetyl-trimethylammonium bromide (CTAB) from the leaf tissues (Doyle and Doyle Citation1987), and the genomic library was constructed via the VAHTS Universal DNA Library Prep Kit for Illumina V3 (Vazyme Biotech Co., Ltd, Nanjing, China). The chloroplast genome of H. distichon was sequenced on the Illumina Novaseq Platform (Illumina, San Diego, CA, USA) at Genepioneer Biotechnologies Inc., Nanjing, China. The high-quality reads (Clean Data) was obtained via fastp (v0.20.0) software (Chen et al. Citation2018), the Clean Data was assembled using SPAdes v3.10.1 (Bankevich et al. Citation2012) and annotated using prodigal v2.6.3 (https://www.github.com/hyattpd/Prodigal), hmmer v3.1b2 (http://www.hmmer.org/)and aragorn v1.2.38 (http://130.235.244.92/ARAGORN/) (Laslett and Canback Citation2004; Hyatt et al. Citation2010; Mistry Citation2013), and manually correct the annotations with another result obtained by blast v2.6 (https://blast.ncbi.nlm.nih.gov/Blast.cgi) (Altschul Citation1990).
The size of the chloroplast genome is 136,462 bp in length, including a large single copy region (LSC) of 80,597 bp, a small single copy region (SSC) of 12,701 bp, and a pair of inverted repeated regions (IR) of 21,582 bp. And the H. distichon chloroplast genome encodes 129 genes, including 83 protein-coding genes, 38 tRNA genes, and eight rRNA genes. The overall GC-content of the chloroplast genome was 38.32%, with the LSC, SSC, and IR regions being 36.31, 32.33, and 43.83%, respectively.
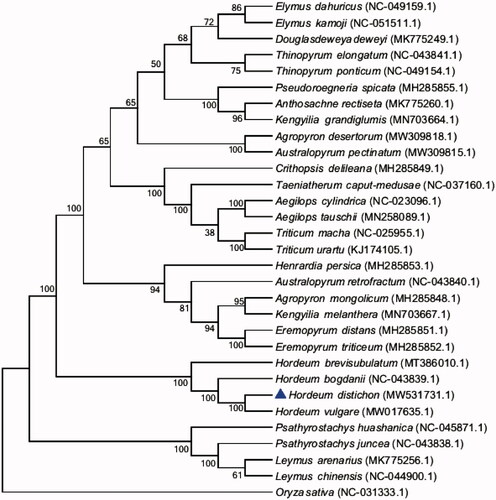
Thirty species in 17 genera of Triticeae and an additional outgroup species (Oryza sativa) were selected from the National Center for Biotechnology Information (NCBI) for investigating the phylogeny of H. distichon (Genbank: MW531731.1). Alignment was conducted using MAFFT software (Katoh and Standley Citation2013). The phylogenetic tree was built using the MEGA 7 software (http://www.megasoftware.net/mega.html) with the maximum likelihood (ML) method and the bootstrap set to 1,000 (Kumar et al. Citation2016; Felsenstein Citation1985). The phylogenetic tree analysis strongly supported that H. distichon was closely relate to Hordeum vulgare (). This study will provide the genome and genetic resources for the further study of H. distichon, and also lay the foundation for the study of chloroplast genome engineering of the H. distichon.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW531731.1; the associated BioProject, SRA, and Bio-Sample numbers are PRJNA743923, SRR15044534, and SAMN20063185, respectively.
Additional information
Funding
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bizuneh WF, Abebe DA. 2019. Malt Barley (Hordeum distichon L.) varieties performance evaluation in North Shewa, Ethiopia. Afr J Agric Res. 14(8):503–508.
- Carpici EB, Celik N. 2012. Correlation and path coefficient analyses of grain yield and yield components in two-rowed of barley (Hordeum vulgare convar. distichon) varieties. Not Sci Biol. 4(2):128–131.
- Chen SF, Zhou YQ, Chen YR, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34(17):i884–890.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 39(4):783–791.
- Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 11:119.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment soft-ware version 7: improvements in performance and usability. Mol BiolE. 30(4):772–780.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32(1):11–16.
- Mistry J, Finn RD, Eddy SR, Bateman A, PuntaM. 2013. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 41(12):e121.
- Niu YF, Li GH, Liu ZY, Zheng C, Liu J. 2020. Sequencing, assembly and annotation of chloroplast genome of Hevea nitida. Non-Wood Forest Res. 38(4):62–71.
- Reichel S, Carvalho DO, Santos JR, Bednar P, Rodrigues JA, Guido LF. 2021. Profiling the volatile carbonyl compounds of barley and malt samples using a low-pressure assisted extraction system. Food Control. 121:107568.
- Schreiber M, Barakate A, Uzrek N, Macaulay M, Sourdille A, Morris J, Hedley PE, Ramsay L, Waugh R. 2019. A highly mutagenised barley (cv. Golden Promise) TILLING population coupled with strategies for screening-by-sequencing. Plant Methods. 15(1):1–14.
- Su XX, Zhao J, Wang ZY. 2020. The complete chloroplast genome of Hordeum brevisubulatum. Mitochondrial DNA B Resour. 5(3):2988–3007.