Abstract
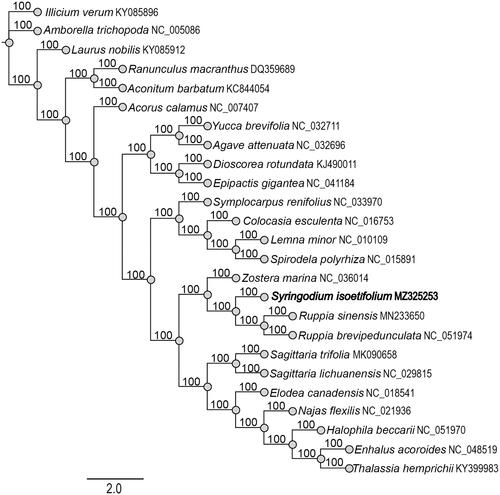
Syringodium isoetifolium (noodle seagrass) is a dioecious perennial seagrass. In this study, the complete chloroplast genome of S. isoetifolium was successfully characterized through next-generation sequencing technology. The cp genome was 159,333 bp in length with a GC content of 35.9%, including LSC (89,055 bp), SSC (19,160 bp), and two IRs (25,559 bp). The genome encoded 131 function genes, including 86 protein-coding genes, 37 tRNA genes, and eight rRNA genes. The phylogenetic analysis indicated that S. isoetifolium was clustered with Zostera and Ruppia.
Syringodium isoetifolium (Asch.) Dandy is a dioecious perennial seagrass in the Cymodoceaceae. It is distributed in Guangdong, Guangxi, Hainan and Taiwan provinces in China (Zheng et al. Citation2013). This species often occurs in mixed seagrass beds with other seagrasses species such as Thalassia hemprichii (Ehrenb. ex Solms) Asch. (Hydrocharitaceae), Cymodocea rotundata Asch. and Schweinf. and C. serrulata (R.Br.) Asch. and Magnus (Cymodoceaceae) (Tomasko et al. Citation1993). Some papers about its screening of microsatellite markers and phylogeography were found (Matsuki et al. Citation2013; Kurokochi et al. Citation2016). However, there are no researches about this species chloroplast genome. In this study, we sequenced, assembled and annotated the complete chloroplast genome of S. isoetifolium with the next-generation sequencing technology to understand its genomic structure and analyze its phylogenetic position.
Samples of Syringodium isoetifolium were collected from Wenchang, Hainan province, China (19.52°E, 110.87°N), and the voucher specimen was deposited in Fourth Institute of Oceanography Herbarium (Shuo Yu, [email protected]) under the voucher number WC201906-1. The total genomic DNA was extracted with the modified CTAB method from cleaned shoots (Yu et al. Citation2020), and sequenced by Illumina Novaseq platform (USA, Illumina company). In total, 4487.1 Mb of raw data and 4389.5 Mb clean data were obtained. The chloroplast genome was assembled with SPAdes v3.9 (Bankevich et al. Citation2012) and gene annotation was performed via PGA software (Qu et al. Citation2019).
The complete chloroplast genome length of Syringodium isoetifolium was 159,333 bp with a GC content of 35.9%. The cp genome had a typical quadripartite structure including one large single-copy region (LSC) (89,055 bp), one small single-copy region (SSC) (19,160 bp), and a pair of inverted repeats (IRs) (25,559 bp). The genome encoded 131 genes including 86 protein-coding genes, 37 tRNA genes, and eight rRNA genes.
To clarify the phylogenetic position of S. isoetifolium, we downloaded 24 completed chloroplast genomes from GenBank database and then aligned them by MATFF v7.308 (Katoh and Standley Citation2013). The phylogenetic tree was constructed with RAxML software (Stamatakis Citation2014) using maximum-like method. Bootstrap values were calculated from 1000 replicates analyses. The phylogenetic tree revealed that S. isoetifolium was clustered with Zostera marina L. (Zosteraceae), Ruppia sinensis Shuo Yu and Hartog and R. brevipedunculata Shuo Yu and Hartog (Ruppiaceae) (). This study was the first time to clearly identify the complete chloroplast of S. isoetifolium and can provide useful information for phylogenetic studies of seagrasses.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The chloroplast genome sequence data that support the findings of this study is deposited in the GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession number MZ325253. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA755496, SRR15498137, and SAMN20826874, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Kurokochi H, Matsuki Y, Nakajima Y, Fortes MD, Uy WH, Campos WL, Nadaoka K, Lian C. 2016. A baseline for the genetic conservation of tropical seagrasses in the western North Pacific under the influence of the Kuroshio Current: the case of Syringodium isoetifolium. Conserv Genet. 17(1):103–110.
- Matsuki Y, Takahashi A, Nakajima Y, Lian C, Fortes MD, Uy WH, Campos WL, Nakaoka M, Nadaoka K. 2013. Development of microsatellite markers in a tropical seagrass syringodium isoetifolium (cymodoceaceae). Conservation Genet Resour. 5(3):715–717.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):1–12.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Tomasko DA, Dawes CJ, Fortes MD, Largo DB, Alava M. 1993. Observations on a multi-species seagrass meadow offshore of Negros Oriental, Republic of the Philippines. Bot Mar. 36:303–312.
- Yu S, Li XJ, Jiang K, Chen XY, Wang XP. 2020. Characteristics of the complete chloroplast genome of Halophila beccarii. Mitochondrial DNA B Resour. 5(1):778–779.
- Zheng FY, Qiu GL, Fan HQ, Zhang W. 2013. Diversity, distribution and conservation of Chinese seagrass species. Biodiv Sci. 21:517–526.