Abstract
Allium hookeri is a rare medicinal plant with unique flavor. In this study, the first complete chloroplast (cp) genome of A. hookeri was sequenced and assembled based on the next generation sequencing. The cp genome is 153,592 bp in length, including a large single-copy (LSC) region of 82,609 bp, a small single-copy (SSC) region of 17,487 bp, and a pair of inverted repeat (IR) regions of 26,748 bp each. The genome encodes 131 genes, including 86 protein-coding genes, 39 tRNA genes, and six rRNA genes. The GC content of whole genome is 36.99%. The phylogenetic analysis based on 24 complete cp sequences revealed that A. hookeri was at the base of the phylogenetic tree, indicating an older species in the Allium genus.
Allium hookeri Thwaites is a member of the genus Allium in Amaryllidaceae. It is a rare plant, narrowly distributed in the 1500–4200 m mountainous areas of Southwest China, Sri Lanka, and Northern India (Xu and Rudolf Citation2000). A. hookeri exhibits high nutritional and medicinal values (Yang et al. Citation2017). It is rich in organosulfur compounds, polyphenols, and allicin (Li et al. Citation2014; Kima et al. Citation2016), which possesses biological activity such as anti-obesity (Park et al. Citation2018; Kim et al. Citation2019), anti-inflammatory (Kim et al. Citation2019; Lee et al. Citation2020), and antimicrobial (Li et al. Citation2014; Kima et al. Citation2016). Consequently, A. hookeri is not only used as a vegetable with unique flavor, but also it has been consumed as medicinal plant. However, only the chloroplast (cp) genome of Allium ferganicum was reported in the genus Allium (Liu et al., Citation2020) . There is no genomic information of A. hookeri that has been reported so far. In this study, the cp genome was sequenced for protecting and excavating the resource of A. hookeri.
Fresh leaves of A. hookeri were collected from Jinfo Mountain, Chongqing, China (107°24′ E, 29°17′ N, 1664 m). The voucher specimen was conserved in Chongqing Institute of Medicinal Plant Cultivation under the accession number of CIMPC-RFM-20210501 (contact person: Fengming Ren, [email protected]). A modified CTAB-based method was used to extract the genomic DNA, and the purity and integrity of the DNA were analyzed by Nanodrop and agarose gel electrophoresis. The genomic DNA was used to generate libraries with insert size of 350 bp and generated about 14 Gb raw reads by Illumina Hiseq 2500 Platform (Illumina, Hayward, CA). The raw data from the platform was removed low-quality reads and adapters by trimmomatic (Bolger et al. Citation2014). Using the clean data with 150 bp paired-end read lengths obtained from the raw data, a cp genome was assembled by NOVOPlasty (Nicolas et al. Citation2016) and annotated by CPGAVAS2 (Shi et al. Citation2019). After manual check and adjustment, the annotated cp genome was submitted to GenBank (MZ557488).
The complete cp genome of A. hookeri was 153,592 bp long and exhibited a typical angiosperm circular cp structure, containing four regions: large single-copy region (LSC: 82,609 bp), small single-copy region (SSC: 17,487 bp), and a pair of inverted repeats (IRs: 26,748 bp). The GC content was 36.99% (whole genome), 34.83% (LSC), 30.01% (SSC), and 42.60% (IR). The GC content of the genome and each genomic region was also typical of angiosperm cp structure. The genome encoded 131 genes, including 86 protein-coding genes, 39 tRNA genes, and six rRNA genes.
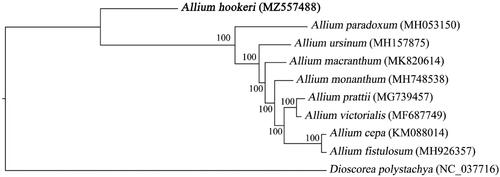
A total of 23 whole genome sequences from the Allium of Amaryllidaceae were downloaded from the GenBank database. The genome sequence of Dioscorea polystachya was used as an outgroup. Finally, the 24 cp genome sequences were multi-aligned by MAFFT software (Katoh and Standley Citation2013). Based on the aligned sequences, a maximum-likelihood phylogenetic tree was built with 1000 bootstrap replicates by IQ-TREE (Nguyen et al., Citation2015)) under parameters of ‘-nt AUTO -m MFP -bb 1000 -bnni’. Phylogenetic analysis showed that A. hookeri was at the base of the phylogenetic tree, which was the oldest species in the selected Allium species ().
Figure 1. Maximum-likelihood phylogenetic tree based on the chloroplast genome sequences of eight Allium (Amaryllidaceae) species and Dioscorea polystachya (outgroup). The GenBank accession numbers is behind the Latin name. The bootstrap support values are beyond each node in the tree. A. hookeri is marked by bold font.
Acknowledgement
We thank Prof. Maoqing Zhou for providing plant material.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MZ557488. The associated BioProject, Bio-Sample, and SRA numbers are PRJNA543381, SAMN20166845, and SRR15098630, respectively.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kima JE, Seo JH, Bae MS, Bae CS, Yoo JC, Bang MA, Cho SS, Park DH. 2016. Antimicrobial constituents from Allium hookeri root. Nat Prod Commun. 11(2):237–238.
- Kim HJ, Lee MJ, Jang JY, Lee SH. 2019. Allium hookeri root extract inhibits adipogenesis by promoting lipolysis in high fat diet-induced obese mice. Nutrients. 11(10):2262.
- Lee SY, Cho SS, Li Y, Bae CS, Park KM, Park DH. 2020. Anti-inflammatory effect of Curcuma longa and Allium hookeri co-treatment via NF-κB and COX-2 pathways. Sci Rep. 10(1):5718.
- Li R, Wang YF, Sun Q, Hu HB. 2014. Chemical composition and antimicrobial activity of the essential oil from Allium hookeri consumed in Xishuangbanna, southwest China. Nat Prod Commun. 9(6):863–864.
- Liu L, Yusupov Z, Suyunkulov H, Jiang Z. 2020. The complete chloroplast genome of Allium ferganicum. Mitochondrial DNA B Resour. 5(3):2772–2773.
- Nguyen L-T, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Nicolas D, Patrick M, Guillaume S. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):4.
- Park S, No K, Lee J. 2018. Anti-obesity effect of Allium hookeri leaf extract in high-fat diet-fed mice. J Med Food. 21(3):254–260.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated n plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Xu J, Rudolf VK. 2000. Allium. In: Wu ZY, Raven PH, editors. Flora of China. Vol. 24. Beijing/St. Louis: Science Press/Missouri Botanical Garden Press; p. 165–202.
- Yang MH, Kim NH, Heo JD, Rho JR, Ock KJ, Shin EC, Jeong EJ. 2017. Comparative evaluation of sulfur compounds contents and antiobesity properties of Allium hookeri prepared by different drying methods. Evid Based Complement Altern Med. 2017:1–10.
