Abstract
Callianthe picta likes a warm and humid climate, is resistant to barrenness, and is easy to reproduce. Its petals and leaves can promote blood circulation and remove blood stasis, and can also be used to relax the muscles and collaterals. In this study, we sequenced the complete chloroplast genome sequence of C. picta to investigate its phylogenetic relationship in the family Abutilon. The complete chloroplast size of C. picta is 160,398 bp, including a large single-copy (LSC) region of 89,088 bp, a small single-copy (SSC) region of 20,138 bp, a pair of invert repeats (IRs) regions of 25,586 bp. The GC content of the whole complete chloroplast genome is 37.0%. We annotated 128 genes in the genome in detail, including 84 protein-coding genes, 36 tRNA genes, and 8 rRNA genes. Phylogenetic analysis indicated that C. picta was closely related to Abutilon theophrati.
Callianthe picta (Gillies ex Hook. & Arn.) Donnell 1834 known as Sida striata is a kind of evergreen shrub of the mallow family Abutilon, mainly native to Brazil, Uruguay and other places in South America, Southwest of China and other parts of the region also have introduced cultivation (Jiang and Jiang Citation2005). Besides being ornamental, petals and leaves of C. picta play a significant role in activating blood circulation and removing blood stasis (Peng et al. Citation2006). The chloroplast genomes are widely used in many research fields such as plant identification and recognition, phylogenetic analysis, and genetic diversity evaluation (Dong et al. Citation2018; Sun et al. Citation2020). In the present study, we reported the chloroplast complete genome sequence of C. picta for the first time and performed phylogenetic analysis to provide valuable materials and information for further in-depth study of this species.
The leaf specimens of C. picta were collected from Kunming, Yunnan, China (102°33′27″E, 25°7′5″N, 1928 m). A specimen was deposited at the Herbarium of Southwest Forestry University (bbg.swfu.edu.cn, Dr.Yao and [email protected]) under the voucher number: SWFU-AAP-APT-3652. Total DNA was extracted using the CTAB method (Doyle and Doyle Citation1987) and sequenced on the lllumina NovaSeq 6000 platform from Annuoyouda Biotechnology Co., Ltd. (Zhejiang, China). About 5.1 GB of high-quality clean reads were generated with adaptors trimmed. Then, the complete chloroplast genome was assembled by GetOrganelle v1.6.2 (Jin et al. Citation2020). Genome annotation was performed by Geneious R9 (Kearse et al. Citation2012) and manually adjusted by comparing it to the reference chloroplast genome of Abutilon theophrasti (GenBank accession number NC053702). The annotated genomic sequence has been submitted to GenBank (accession number: MZ615336).
The chloroplast genome of C. picta is a circular DNA molecule with a length of 160,398bp, which contains a large single-copy region (LSC), a small single-copy region (SSC), and a pair of inverted repeat sequences (IRs), with lengths of 89,088, 20,138, and 25,586 bp, respectively. The content of guanine (G) and cytosine (C) in the whole chloroplast genome is 37.0%. In addition, it contains 128 genes with different functional classifications, of which 84 are functional genes encoding proteins, 36 are tRNA genes, and 8 are rRNA genes.
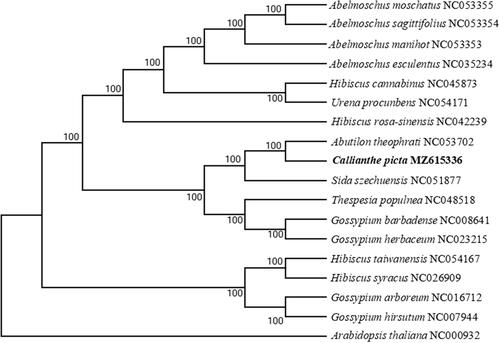
Available chloroplast genomic information of Abutilon plants was limited. So we selected 16 related plant species to study their phylogenetic position with C. picta. First, we used the Clustal W program to compare multiple sequences with default parameters, saved the comparison results, and imported them into MEGA X. We used the neighbor-joining (NJ) method to construct the evolutionary tree, with the combination of the boot Tstrap method (1000 replicates) (Kumar et al. Citation2018). Arabidopsis thaliana (NC000932) was served as the out-group. It was found that C. picta was more closely related to A. theophrati than to species from other taxonomic groups (). The complete genome-wide determination of C. picta chloroplasts provides new molecular data for elucidating the evolutionary relationship of Malvaceae and other biological studies.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The voucher specimens of Callianthe picta were deposited at the Herbarium of Southwest Forestry University, Kunming, Yunnan, China (accession number: SWFU-AAP-APT-3652). The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/) under the accession no. MZ615336. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA481636, SRR7589393, and SAMN09691266, respectively.
Additional information
Funding
References
- Dong W, Xu C, Wu p, Cheng T, Yu J, Zhou S, Hong DY. 2018. Resolving the systematic positions of enigmatic taxa: manipulating the chloroplast genome data of Saxifragales. Mol Phylogenet Evol. 126:321–330.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19(1):11–15.
- Jiang YH, Jiang XL. 2005. Extraction and physicochemical properties of pigment from lantern flower. Jiangsu Agric Sci. 6:125–127.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Phylogenet Evol. 35(6):1547–1549.
- Peng SC, Liu FN, Liu LR. 2006. Golden bells swaying in the wind. Chinese Flower Bonsai. 2:15.
- Sun J, Wang Y, Liu Y, Xu C, Yuan Q, Guo L, Huang L. 2020. Evolutionary and phylogenetic aspects of the chloroplast genome of Chaenomeles species. Sci Rep. 10(1):11466.