Abstract
This study presents the first complete mitochondrial genome of the Hipposideros pendleburyi (Pendlebury's leaf-nosed bat), an endemic species in Thailand. The mitochondrial genome was 16,820 bp in length and contains 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNA genes, and a control region. The overall base composition was 31.5% A, 26.2% T, 28.3% C, and 14.0% G. A maximum-likelihood tree revealed that H. pendleburyi was grouped with Hipposideros armiger within the Hipposideridae clade, which has Rhinolophidae as a sister clade.
The Pendlebury's leaf-nosed bat (Hipposideros pendleburyi) was named by Chasen in 1936 but had long been included in Hipposideros turpis (Lekagul and McNeely Citation1977; Corbet and Hill Citation1992; Francis Citation2008). Only a decade ago, it has been regarded as a distinct species and known only from peninsular Thailand (Soisook Citation2011; Thong et al. Citation2012). This species was found in limestone areas of seven provinces in peninsular Thailand with a colony size of up to 800 individuals (Soisook Citation2019). It has been assessed as a vulnerable species on the Red List of Threatened Species by IUCN, and its population size has continuously declined due to habitat disturbance by human activities and limestone quarrying (Soisook Citation2019). Until now, there is only one complete mitochondrial genome (Hipposideros armiger) of the bat in the family Hipposideridae available (Dong et al. Citation2017). The complete mitochondrial genome of H. pendleburyi provides valuable information for inferring the phylogenetic relationships of Chiroptera order and a foundation for future research.
A male specimen of H. pendleburyi was collected from Tham Le Stegodon Cave, Palian District, Trang Province, Thailand (7.141 N, 99.789 E). Bat sampling in this study was permitted by the Department of National Park, Wildlife and Plant Conservation (project number 6210306). Collection and handling of bats followed the guidelines of the American Society of Mammalogists (Sikes Citation2016). The specimen was deposited in the Mammal Collection of the Princess Maha Chakri Sirindhorn Natural History Museum, Prince of Songkla University (PSU), Hat Yai, Songkhla, Thailand (http://www.biology.sci.psu.ac.th/pipat-soisook/, Pipat Soisook: [email protected]) under the voucher number PSUZC-MM.2021.6. Total DNA was extracted using QIAamp Tissue Kit (Qiagen, Germany). A DNA sequencing library was constructed and paired-end reads (150 bp) were sequenced by Illumina HiSeqX Ten sequencer (Illumina, Singapore).
The H. pendleburyi mitochondrial genome was assembled de novo using MitoZ 2.4 (Meng et al. Citation2019) and annotated using the MITOS web server (Bernt et al. Citation2013). Protein-coding genes (PCGs) and RNA genes were confirmed using the Basic Local Alignment Search Tool (BLAST) (Altschul et al. Citation1990). The complete mitochondrial genome (GenBank Accession Number: MZ196220.1) was 16,820 bp in length including 13 PCGs, 2 ribosomal RNA genes (rRNAs), 22 transfer RNA genes (tRNAs), and a non-coding control region. The overall base composition was 31.5% A, 26.2% T, 28.3% C, and 14.0% G. The PCGs utilized the standard mitochondrial start codon ATN (10 with ATG, 3 with ATA) and the regular stop codons (TAA or TAG) except for cytb (AGA). The incomplete stop codon was observed in Nd4 (T–) and coxIII (TA–).
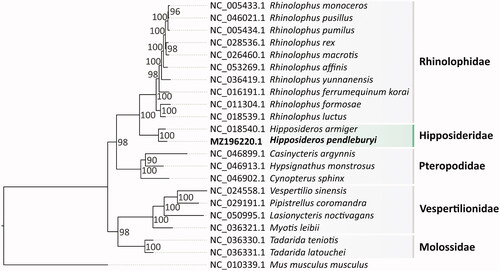
Phylogenetic analysis of the H. pendleburyi mitogenome was performed based on 13 PCGs from 21 Chiroptera species and one outgroup (Mus musculus musculus). We concatenated sequences from 13 PCGs and performed multiple alignments using MUSCLE (Edgar Citation2004). Subsequently, amino acid replacement models were estimated using ModelTest-NG (Darriba et al. Citation2020) and a maximum-likelihood phylogenetic tree was constructed by RAxML-NG (Kozlov et al. Citation2019) with 1000 bootstrap replicates (). The result showed that H. pendleburyi was well grouped with H. armiger within the Hipposideridae clade, which had Rhinolophidae as a sister clade. Our result is consistent with the previous study by Lei and Dong (Citation2016).
Acknowledgments
We are grateful to the National Science and Technology Development Agency (NSTDA), Thailand for financial support. We thank Awatsaya Pimsai and Sakiya Morlor at Prince of Songkla University (PSU) for help with fieldwork and sample collection.
Disclosure statement
The authors declare no competing financial interests.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MZ196220.1. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA763996, SRR15927285, and SAMN21465651 respectively.
Additional information
Funding
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Corbet GB, Hill JE. 1992. The mammals of the Indomalayan Region. Oxford: Natural History Museum and OUP Press. p. 488.
- Darriba D, Posada D, Kozlov AM, Stamatakis A, Morel B, Flouri T. 2020. ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Mol Biol Evol. 37(1):291–294.
- Dong D, Lei M, Hua P, Pan YH, Mu S, Zheng G, Pang E, Lin K, Zhang S. 2017. The genomes of two bat species with long constant frequency echolocation calls. Mol Biol Evol. 34(1):20–34.
- Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 5:113.
- Francis CM. 2008. Mammals of Thailand and South-East Asia. Bangkok: Asia Books. p. 392.
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. 2019. RAxML-NG: a fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 35(21):4453–4455.
- Lei M, Dong D. 2016. Phylogenomic analyses of bat subordinal relationships based on transcriptome data. Sci Rep. 6:27726.
- Lekagul B, McNeely JA. 1977. Mammals of Thailand. Association for the Conservation Wildlife, Bangkok. 758 pp.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for ANIMAL mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63–e63.
- Sikes RS. 2016. 2016 Guidelines of the American Society of Mammalogists for the use of wild mammals in research and education. J Mammal. 97 (3):663–688.
- Soisook D. 2011. A checklist of bats (Mammalia: Chiroptera) in Thailand. J Wildlife Thailand. 18:121–151.
- Soisook P. 2019. Hipposideros pendlebury. The IUCN Red List of Threatened Species 2019: e.T80224655A95642195. https://doi.org/http://dx.doi.org/10.2305/IUCN.UK.2019-3.RLTS.T80224655A95642195.en
- Thong V, Puechmaille S, Denzinger A, Bates P, Dietz C, Csorba G, Soisook P, Teeling E, Matsumura S, Furey N, et al. 2012. Systematics of the Hipposideros turpis complex and a description of a new subspecies from Vietnam. Mammal Review. 42(2):166–192.