Abstract
The first complete mitochondrial genome of Lixus subtilis Boheman is reported in this study. The circular genome is 15,223 bp long, including a standard set of 21 transfer RNAs (tRNAs), 2 ribosomal RNAs (rRNAs), 13 protein-coding genes, and a non-coding control region. The trnI gene was not found in the L. subtilis mitogenome. All tRNAs had the typical cloverleaf structure, except for trnS1, which lacked the dihydrouridine arm. The phylogenetic tree of 13 Curculionidae species based on the concatenated nucleotide sequences of complete mitochondrial genomes strongly supported that L. subtilis is closely related to Curculioninae and Molytinae.
Lixus subtilis Boheman, 1835 (Coleoptera: Curculionidae) is widely distributed (Volovniket al. Citation2015; Davidian et al. Citation2017) and a primary pest of beets, amaranth, and gray vegetables (Davidian et al. Citation2017). In recent years, L. subtilis has had outbreaks in the quinoa growing areas of Shanxi and Beijing, China, causing large crop losses (Zhang et al. Citation2017). 30 adult specimens of L. subtilis were collected from the quinoa field in Supo Village, Supo Township, Jingle County, Shanxi Province, China (112.2°E, 38.4°N) on 10 May 2018. One specimen was deposited in the College of Plant Protection, Shanxi Agricultural University, Taiyuan, China (Kun Xing, [email protected]) under the accession no. YFMgnjP2018393.
Thirty specimens were used in the mitogenomic studies. The complete mitogenome of L. subtilis is a representative circular DNA molecule with a length of 15,223 bp (GenBank accession no. MW413392). Thirteen protein-coding genes (PCGs), 21 transfer RNA genes (tRNAs), the large and small ribosomal RNA unit genes (rrnL and rrnS), and a large non-coding region (putative control region) were contained by this mitogenome. In Coleoptera, the order and orientation of the mitochondrial genomes have been retained from the ancestral gene order, apart for the tRNA gene, which may be deleted or rearranged in some species (Timmermans and Vogler Citation2012). The trnI was not found in the L. subtilis mitogenome, as observed in Eucryptorrhynchus chinensis (Oliver, 1790) and Naupactus xanthographus (Germar, 1824) (Tang et al. Citation2017; Yang et al. Citation2018). The nucleotide composition of L. subtilis was significantly biased: A, G, C, and T accounted for 40.2%, 9.6%, 14.7%, and 35.5%, respectively; A + T contents totaled 75.7%. In this genome, the GC-skew and AT-skew were −0.209 and 0.063, respectively. Gene overlaps had a total of 45 bp and were present in ten gene junctions. The largest gene overlap (−17 bp) was present between trnF and nad5, in which intergenic spacers totaling 53 bp appeared in 11 positions and ranged in size from 1 to 18 bp. The control region had A + T content of 72.0% with 571 bp in length and was present between the rrnS and trnQ genes.
It was predicted that all 21 tRNAs had typical cloverleaf secondary structures, but the gene trnS1 lacked a stable DHU arm. This result was as the same as those reported in other insect mitogenomes (Yuan et al. Citation2016). The rrnL gene was located between the trnL1 and trnV genes, and the trnV and rrnS genes were located between the trnV gene and the control region. The rrnL gene had an A + T content of 81.3% and a length of 1291 bp. The rrnS gene was 813 bp in length and had A + T content of 76.3%. Eleven PCGs had a typical ATN codon. PCGs nad2, nad4, nad4l, and cob started with ATG; cox1, cox2, atp8, cox3, and nad6 started with ATT; and atp6 and nad3 started with ATA. However, nad5 and nad1 started with GTG and TTG, respectively. Ten PCGs terminated with TAA and two terminated with TAG (atp8 and nad1), whereas one terminated with an incomplete stop codon TA (nad4).
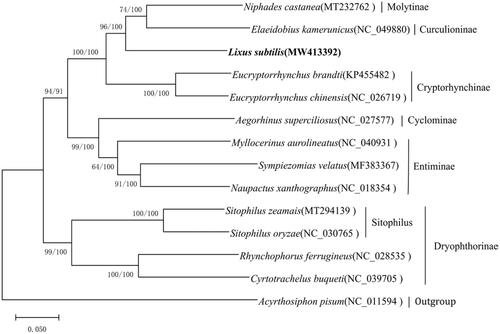
For the phylogenetic analysis, the nucleotide sequences of complete mitochondrial genomes from 13 species (Curculionidae) and outgroups from Acyrthosiphon pisum (Harris, 1776) (Hemiptera, Aphididae) were used. This phylogenetic analysis was performed using the maximum likelihood (ML) method with 1,000 bootstrap replicates using MEGA-X and via Bayesian inference (BI) using MrBayes (Ronquist and Huelsenbeck Citation2003; Kumar et al. Citation2018). There was strong support for clustering of L. subtilis with Curculioninae and Molytinae (), indicating that L. subtilis is more closely related to Curculioninae and Molytinae than other subfamilies.
Disclosure statement
The authors report no conflicts of interests. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/genbank under the accession no. MW413392. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA757215, SRR15603403, and SAMN20955048, respectively.
Additional information
Funding
References
- Davidian GE, Korotyaev BA, Gültekin L. 2017. On the distribution of the weevil Lixus subtilis Boheman, 1835 (Coleoptera, Curculionidae: Lixinae). Entmol Rev. 97(5):594–601.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Tang PA, Zhang L, Li XP, Li FF, Yuan ML. 2017. The complete mitochondrial genome of Sympiezomias velatus (Coleoptera: Curculionidae). Mitochondrial DNA B Resour. 2(2):449–450.
- Timmermans M, Vogler A. 2012. Phylogenetically informative rearrangements in mitochondrial genomes of Coleoptera, and monophyly of aquatic elateriform beetles (Dryopoidea). Mol Phylogenet Evol. 63(2):299–304.
- Volovnik SV, Nazarenko VY, Sheshurak PN. 2015. Weevils of the subfamily Lixinae (Coleoptera: Curculionidae) of Chernigov Province (Ukraine): species composition and geographic distribution. Ukrainskii Entomologichnii Zhurnal. 1(2):76–83.
- Yang WJ, Yang DX, Xu KK, Cao Y, Meng YL, Wu Y, Li GY, Zhang GZ, Wang YW, Li C. 2018. Complete mitochondrial genome of the bamboo snout beetle, Cyrotrachelus buqueti (Coleoptera: Curculionidae). Mitochondrial DNA B Resour. 3(1):88–89.
- Yuan ML, Zhang QL, Zhang L, Guo ZL, Liu YJ, Shen YY, Shao RF. 2016. High-level phylogeny of the Coleoptera inferred with mitochondrial genome sequences. Mol Phylogenet Evol. 104:99–111.
- Zhang GF, Zhang JL, Wan FH, Mei L, Xian XQ, Dong J, Guo ZJ. 2017. Outbreak of Lixus subtilis Boheman on quinoa. Plant Protection. 43(2):202–207.