Abstract
Leucomeris decora is a traditional medicinal plant that is listed as an endangered species in China. Recently, L. decora has become locally rare. Here the complete chloroplast genome of L. decora was assembled and reported for the first time. Its plastome was 151,491 bp in length, including a large single-copy region (LSC; 83,155 bp), a small single-copy region (SSC; 18,216 bp), and a pair of inverted repeated regions (IRa and IRb; 25,060 bp). The overall GC content was 37.8%, and the genome contains 134 genes, including 92 protein-coding genes, 8 rRNA genes, and 34 tRNA genes. Phylogenetic analysis of thirteen representative species from the family of Asteraceae showed that L. decora is clustered into one clade with Gerbera jamesonii with high bootstrap values, indicating a close relationship between these two species.
Leucomeris decora Kurz belongs to the family Asteraceae, which is generally distributed in Southwest China (Funk et al. Citation2009; Zhao and Gong Citation2012). The plant’s leaves and stems have been used as a traditional anti-inflammatory medicine (Liu et al. Citation2018). Due to rapid habitat fragmentation and unrestricted utilization, L. decora has become endangered with a drastically reduced population size, and it is now listed in the IUCN Red List of Threatened Species and the Red Book of Chinese Plants (Fu and Jin Citation1992).
There is no clear morphological divergence between L. decora and other woody species of Asteraceae, such as Nouelia insignis Franch. The bisexual corolla of Leucomeris is radially symmetrical, and the bisexual corolla of Nouelia is symmetrical on both sides (Peng et al. Citation2002). Both of these genera possess the same number of chromosomes with 2n = 54, which differs from that in other genera of Asteraceae (Panero and Funk Citation2008; Zhao and Gong Citation2015). Despite differences in their floral traits, the genera of Leucomeris and Nouelia are clustered into a single group based on morphological and molecular evidence (chloroplast DNA and nuclear ribosomal DNA) (Zhao and Gong Citation2015). Although the taxonomic status of the genus Leucomeris has been elucidated to some extent, few studies have focused on the phylogenetic and adaptive evolution of this genus at the plastomic level.
Fresh leaves of L. decora were collected from Yunnan, China (N 24.943°, E 98.868°), and a specimen of L. decora (specimen No. 622) was deposited at the Herbarium, Kunming Institute of Botany, Chinese Academy of Sciences (under the voucher No. 622, Dr. Zhao, [email protected]). Total genomic DNA was extracted from leaves using the modified CTAB method (Suzana et al. Citation2015) and NanoDrop 2000 spectrophotometer was used for determined the concentration of DNA. The sequenced library was constructed and generated with a paired-end library of 2 × 250 bp using Illumina HiSeq X platform (Illumina, Inc., America) with the modified manuscript. The genomic DNA were sequenced more than 6 GB raw data through the Illumina platform at Novogene (Beijing, China). All raw reads were trimmed and assembled with the GetOrganelle software (Jin et al. Citation2020). The chloroplast genome was annotated with both PGA and GB2sequin (https://chlorobox.mpimp-golm.mpg.de/geseq.html) using the chloroplast genome of Gerbera jamesonii Adlam (Genbank accession No. MN087227) as the reference. The tRNAscan-SE program was employed to identify tRNA genes with default parameters. A gene map of the annotated L. decora genome was drawn using the online tool OGdraw (https://chlorobox.mpimp-golm.mpg.de/geseq.html). Finally, the whole chloroplast genome of L. decora was submitted to GenBank under the accession No. MT386593.
The chloroplast genome of L. decora is 151,491 bp in length and exhibits a typical quadripartite structure: the two inverted repeat (IR) regions of 25,060 bp were separated by a large single-copy region (LSC; 83,155 bp) and a small single-copy (SSC; 18,216 bp) region. The GC content of the chloroplast genome was 37.8%, and the GC content of the LSC, SSC, and IR regions were 35.4, 31.5, and 43.2%, respectively. Ninety-two protein-coding genes, 34 tRNAs, and 8 rRNAs were detected in the whole chloroplast genome sequence. Out of these 134 genes, there were 20 duplicate genes (9 protein-coding genes, 7 tRNAs, and 4 rRNAs) in the IR regions and 20 genes which contained introns.
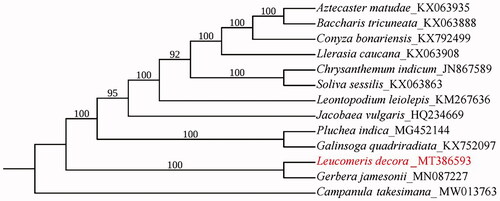
The whole chloroplast genome sequences of 13 species were downloaded to analyze the phylogenetic relationship among L. decora and other species of Asteraceae. Campanula takesimana Nakai (Genbank accession No. MW013763) in the family Campanulaceae was used as the outgroup. Alignment was conducted using MAFFT (Katoh and Standley Citation2013). Phylogenetic analysis was carried out based on maximum likelihood analysis by IQ-TREE 1.5.5 software (Nguyen et al. Citation2015), with TVM + F+R2 as the best nucleotide substitution model and 10,000 bootstrap replicates. The results of phylogenetic analysis showed that L. decora was close to Gerbera jamesonii Bolus, as reported by Fu et al. (Citation2016), which supports that the chloroplast genome of L. decora provides useful genetic information for further studies on the genetic diversity and conservation of Asteraceae species ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] under the accession no. MT386593. The associated Bioproject, SRA, and Bio-Sample numbers are PRJNA729624, SRR14520563, and SAMN19136515, respectively.
Additional information
Funding
References
- Fu LG, Jin JM. 1992. The red book of Chinese plants: rare and Endangered plants. Beijing: Science Press.
- Fu ZX, Jiao BH, Nie B, Zhang GJ, Gao TG, China Phylogeny Consortium. 2016. A comprehensive generic-level phylogeny of the sunflower family: implications for the systematics of Chinese Asteraceae. J Sytemat Evol. 54(4):416–437.
- Funk VA, Susanna A, Steussy TF, Bayer RJ. 2009. Systematics, evolution, and biogeography of Compositae. Vienna, Austria: International Association for Plant Taxonomy.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis C, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241–271.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Liu L, Liu YF, Li BC, Zhang M. 2018. Study on in vitro anti-inflammatory activity of methanol extracts from the barks, branches and leaves and chemical constituent of Gochnatia decora (Chinese). J Plant Resour Environ. 27:101–103.
- Nguyen LT, Schmidt HA, Haeseler AV, Minh BQ. 2015. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Panero JL, Funk VA. 2008. The value of sampling anomalous taxa in phylogenetic studies: major clades of the Asteraceae revealed. Mol Phylogenet Evol. 47(2):757–782.
- Peng YL, Sun H, Gu ZJ. 2002. Cytological study on Nouelia and Leucomeris (Compositae) (Chinese). Plant Diversity. 24:82–86.
- Suzana M, Rahimah AR, Maizura I, Singh R. 2015. A simple and rapid protocol for isolation of genomic DNA from oil palm leaf tissue. J Oil Palm Res. 27:282–287.
- Zhao YJ, Gong X. 2015. Genetic divergence and phylogeographic history of two closely related species (Leucomeris decora and Nouelia insignis) across the “Tanaka Line” in Southwest China. BMC Evol Biol. 15(1):1–13.
- Zhao YJ, Gong X. 2012. Genetic structure of the endangered Leucomeris decora (Asteraceae) in China inferred from chloroplast and nuclear DNA markers. Conserv Genet. 13(1):271–281.