Abstract
The complete mitochondrial genome of Haemadipsa tianmushana Song 1977 from China has been determined and reported for the first time in this study. It was 14,625 bp in length and consisted of 22 tRNA genes, 2 rRNA genes, 13 protein-coding genes (PCGs), and 3 control regions. The nucleotide base content of the complete mitogenome for this species was 35.1% A, 10.5% C, 11.6% G, and 42.8% T. The tRNA genes were ranged from 57 bp (SerTCT) to 66 bp (GlnTTG) in length. The phylogenetic analyses indicated that Hirudinea is a mono-phyletic clade. And it includes Whitmania acranulata, Whitmania pigra, Whitmania laevis, Zeylanicobdella arugamensis, Ozobranchus jantseanus and Placobdella lamothei. In Hirudiniformes, H. tianmushana and three species of Haemopidae were obviously clustered into two independent branches. This result is consistent with a taxonomy that they all belong to the same suborder. This study adds to the genetic resources currently available for the species.
Haemadipsa tianmushana Song 1977, belongs to the family Haemadipsidae, suborder Hirudiniformes (Schoch et al. Citation2020). Land leeches in Haemadipsidae are mostly from the humid tropical rainforest habitats and habitually take blood from humans and other animals (Won et al. Citation2014; Huang et al. Citation2019). Compared with most blood‐feeding arthropods, they have the larger gut capacity, being highly abundant and easy to collect, therefore haemadipsid leeches may be particularly useful as iDNA sources for biodiversity monitoring. Drinkwater identified sequences from 14 mammalian genera, spanning nine families and five orders using Haemadipsa picta and Haemadipsa sumatrana. (Drinkwater et al. Citation2019). H. tianmushana is endemic leech species only distributed in China, which might serve as suitable and useful samples of biodiversity research. In addition, compared with nuclear DNA, the mitochondrial DNA of many animals tends to relatively higher mutation rate (Brown et al. Citation1979; Duda Citation2021). Almost all the mitogenome sequence datasets included the Mitochondrial Cytochrome C Oxidase I (COI) sequence. COI sequences variation among the clariid fishes of India (Clarias magur, Clarias dussumieri, and Clarias gariepinus) and their relationship with other representative clariids (Devassy et al. Citation2016). Therefore mitochondrial DNA is considered one of the most reliable molecular markers for phylogenetic studies (Shen et al. Citation2011; Chen et al. Citation2012; Xu et al. Citation2014; Vella and Vella Citation2021).
Specimen samples were obtained from Mount Huangshan, Anhui Province, China (118°1′E, 30°1′N). A specimen of H. tianmushana was deposited at the Institute of Medicinal Plant Development(Linchun Shi, [email protected]) under the voucher number HS22. The genomic DNA was extracted from the tissue sample using TIANamp Genomic DNA Kit (Tiangen, Beijing, China) following the manufacturer’s protocol. A NanoDrop 2000 ultra-micro spectrophotometer (Thermo Scientific, USA) was used to measure the yield and purity of genomic DNA, and next quantified using Qubit 4.0 (Thermo Scientific, USA). This process was followed by creating PCR-free libraries from the extracted genomic DNA (Liu et al. Citation2021). Next-generation sequencing was performed using the Illumina Novaseq platform. Trimmomatric v0.38 was employed to filter the low-quality reads and the sequencing adapter (Bolger et al. Citation2014). The complete circular mitogenome was de novo assembled using GetOrganelle v1.7.4 (Li et al. Citation2020). The mitogenome was annotated using MITOS (Bernt et al., Citation2013). To confirm the exactness of the mitochondrial genome, the manual correction was performed by annelid multiple sequence alignment. The mitogenomic sequencing data were individually mapped to the COI databases using BOWTIE2 v2.4.4 (Langmead and Salzberg Citation2012). PCGs were identified by ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/) using mitochondrial genetic codes.
The whole mitochondrial genome of H. tianmushana is a circular molecule and 14,625 bp in length. It was deposited in GenBank under accession no. MZ189977. It consists of 22 tRNA genes, 2 rRNA genes, 13 protein-coding genes (PCGs), and 3 control regions (Bronstein et al. Citation2018). The nucleotide base content of H. tianmushana mitogenome was 35.1% A, 10.5% C, 11.6% G, and 42.8% T. The 22.1% of (G + C) showed great preference to AT. It follows the typical gene order and composition of fish species (Satoh et al. Citation2016). The mitogenome of H. tianmushana consists of 13 PCGs, PCGs’ lengths range between 171 bp (atp8) and 1722 bp (nad5). Most of the PCGs utilize ATG as their termination codon except nad5, nad6, cox1, and nad1 similar to the structure and variability of most other fish mitogenomes (Satoh et al. Citation2016). The mitogenome of H. tianmushana contains 22 tRNA genes varying from 57 bp (SerTCT) to 66 bp (GlnTTG) in length, and all produced the expected typical cloverleaf structure. There are three control regions. The first control region is 49 bp in length located between tRNA-Trp and tRNA-Arg. The second is 85 bp in length located between tRNA-Arg and atp6. And the last is 124 bp in length located between nad4 and tRNA-Met.
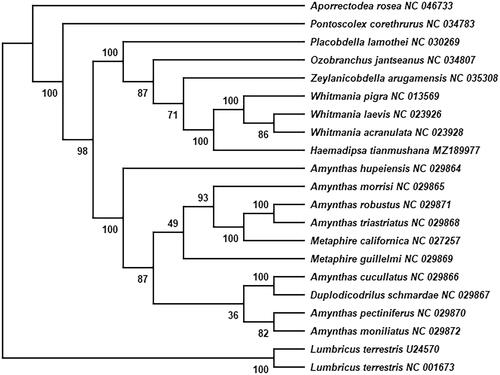
The mitogenome of H. tianmushana was aligned with other 18 Clitellata species whose mitogenomic protein sequences have more than 60% sequence homology with that of H.tianmushana using ClustalW (Thompson et al. Citation1994). The phylogenetic position of H. tianmushana was confirmed using RAxML v8.0.0 (Stamatakis Citation2014) to create a Maximum-Likelihood (ML) tree with GTRGAMMA model. Lumbricus terrestris (U24570 and NC001673) was used as an outgroup (Liu et al. Citation2017). The result () suggests that Hirudinea is a mono-phyletic group. And it includes Whitmania acranulata, Whitmania pigra, Whitmania Laevis, Zeylanicobdella arugamensis, Ozobranchus jantseanus, and Placobdella lamothei. In Hirudiniformes, H.tianmushana and three species of Haemopidae were obviously clustered into two independent branches. The phylogenetic analysis results were consistent with the morphological observations. This study provides reliable mitogenome information from H. tianmushana that could facilitate the species identification and phylogenetic study of H. tianmushana.
Disclosure statement
No potential conflict of interest was reported by the authors(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in Genbank of NCBI at https://www.ncbi.nlm.nih.gov/under the accession no. MZ189977. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA743159, SRR15020542, and SAMN20000658, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Bronstein O, Kroh A, Haring E. 2018. Mind the gap! The mitochondrial control region and its power as a phylogenetic marker in echinoids. BMC Evol Biol. 18(1):80.
- Brown WM, George M, Wilson AC. 1979. Rapid evolution of animal mitochondrial DNA. Proc Nat Acad Sci USA. 76(4):1967–1971.
- Chen D-X, Chu W-Y, Liu X-L, Nong X-X, Li Y-L, Du S-J, Zhang J-S. 2012. Phylogenetic studies of three sinipercid fishes (Perciformes: Sinipercidae) based on complete mitochondrial DNA sequences. Mitochondrial DNA. 23(2):70–76.
- Devassy A, Kumar R, Shajitha PP, John R, Padmakumar KG, Basheer VS, Gopalakrishnan A, Mathew L. 2016. Genetic identification and phylogenetic relationships of Indian clariids based on mitochondrial COI sequences. Mitochondrial DNA A DNA Mapp Seq Anal. 27(5):3777–3780.
- Drinkwater R, Schnell IB, Bohmann K, Bernard H, Veron G, Clare E, Gilbert MTP, Rossiter SJ. 2019. Using metabarcoding to compare the suitability of two blood-feeding leech species for sampling mammalian diversity in North Borneo. Mol Ecol Resour. 19(1):105–117.
- Duda TF. 2021. Patterns of variation of mutation rates of mitochondrial and nuclear genes of gastropods. BMC Ecol Evo. 21(1):13.
- Huang T, Liu Z, Gong X, Wu T, Liu H, Deng J, Zhang Y, Peng Q, Zhang L, Liu Z, et al. 2019. Vampire in the darkness: a new genus and species of land leech exclusively bloodsucking cave-dwelling bats from China (Hirudinda: Arhynchobdellida: Haemadipsidae). Zootaxa. 4560(2):272
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9 (4):357–359.
- Li X, Lin C-Y, Yang J-B, Yu W-B. 2020. De novo assembling a complete mitochondrial genome of Pedicularis rex (Orobanchaceae) using GetOrganelle toolkit. Mitochondrial DNA B Resour. 5(1):1056–1057.
- Liu J, Mu W, Shi M, Zhao Q, Kong W, Xie H, Shi L. 2021. The species identification in traditional herbal patent medicine, Wuhu San, based on shotgun metabarcoding. Front Pharmacol. 12:12.
- Liu X, Luo D, Zhao Y, Zhang Q, Zhang J. 2017. Complete mithochondrial genome of Ozobranchus jantseanus (Hirudinida: Arhychobdellida: Ozobranchidae)). Mitochondrial DNA B Resour. 2(1):232–233.
- Satoh TP, Miya M, Mabuchi K, Nishida M. 2016. Structure and variation of the mitochondrial genome of fishes. BMC Genom. 17(1):719.
- Schoch CL, Ciufo S, Domrachev M, Hotton CL, Kannan S, Khovanskaya R, Leipe D, Mcveigh R, O’Neill K, Robbertse B, et al. 2020. NCBI taxonomy: a comprehensive update on curation, resources and tools. Database. 2020:baaa062.
- Shen X, Wu Z, Sun M, Ren J, Liu B. 2011. The complete mitochondrial genome sequence of Whitmania pigra (Annelida, Hirudinea): The first representative from the class Hirudinea. Comp Biochem Physiol Part D Genomics Proteomics. 6(2):133–138.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22(22):4673–4680.
- Vella N, Vella A. 2021. Characterization of the complete mitogenome of Haifa grouper, Hyporthodus haifensis (Perciformes: Serranidae), and its phylogenetic position within Epinephelini. Mitochondrial DNA B Resour. 6(4):1287–1289.
- Won S, Park BK, Kim BJ, Kim HW, Kang JG, Park TS, Seo HY, Eun Y, Kim KG, Chae JS, et al. 2014. Molecular identification of Haemadipsa rjukjuana (Hirudiniformes: Haemadipsidae) in Gageo Island, Korea. Korean J Parasitol. 52(2):169–175.
- Xu Y, Nie J, Hou J, Xiao L, Lv P. 2014. Complete mitochondrial genome of Hirudo nipponia (Annelida, Hirudinea). Mitochondrial DNA A DNA Mapp Seq Anal. 27(1):257–258.