Abstract
We have sequenced mitochondrial genome of Uroleucon erigeronense (Thomas, 1878) isolated from Erigeron canadensis in Korea. The circular mitogenome of U. erigeronense is 15,691 bp long including 13 protein-coding genes, two ribosomal RNA genes, 22 transfer RNAs, and a single control region of 968 bp. AT ratio is 84.2%. Additional phylogenetic studies of aphid mitogenomes are required due to the inconsistency found in the three phylogenetic trees.
The genus Uroleucon Mordvilko, 1914 (Hemiptera: Aphididae) consisting of more than 200 species is distributed world-widely (Nafría et al. Citation2007). Uroleucon erigeronense (Thomas, 1878) belonging to the subgenus Lambersius Olive, 1965, which is one of five subgenera of Uroleucon, was introduced to Europe (Blackman and Eastop Citation2006) and Australia recently (Brumley and Watson Citation2017). U. erigeronense usually feeds on the Erigeron species; but its host range was revealed widely including another Asteraceae plant species, such as Dieteria canescens (Pursh) A.Gray and Ericameria nauseosa (Pall. ex Pursh) G.L.Nesom & G.I.Baird (Jensen et al. Citation2010). In Korea, U. erigeronense is already considered a native species (Park, An, et al. Citation2020). Here, we presented the complete mitogenome of U. erigeronense isolated from Erigeron canadensis in Korea as the first mitogenome of Uroleucon genus.
Like the previous studies that complete organelle genomes were rescued from the sample contains multiple organisms (Bae et al. Citation2020; Park, Xi, Park, Lee Citation2020; Park, Xi, Park, Nam, et al. Citation2020; Choi et al. Citation2021; Park, Lee, et al. Citation2021; Park, Xi, Kim, et al. Citation2021; Park, Xi, Park Citation2021), we sequenced the DNA (37.529708 N, 126.842867E; InfoBoss Cyber Herbarium (IN); IB-30034; Contact person: Jongsun Park, [email protected]) prepared from the E. canadensis sample with U. erigeronense extracted using DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Raw sequences obtained from Illumina NovaSeq6000 (Macrogen Inc., South Korea) were filtered by Trimmomatic v0.33 (Bolger et al. Citation2014), de novo assembled by Velvet v1.2.10 (Zerbino and Birney Citation2008). Gaps were closed with GapCloser v1.12 (Zhao et al. Citation2011), BWA v0.7.17, and SAMtools v1.9 (Li et al. Citation2009; Li Citation2013) in the environment of Genome Information System (GeIS; http://geis.infoboss.co.kr/), which utilized in previous insect mitogenome studies (Han et al. Citation2017; Park, Jung, et al. Citation2019; Park, Park, et al. Citation2019; Lee et al. Citation2020; Park, Xi, et al. Citation2020; Jung et al. Citation2021; Park and Park Citation2021). Geneious Prime® 2020.2.4 (Biomatters Ltd, Auckland, New Zealand) was used to annotate mitogenome based on Sitobion avenae (Fabricius, 1775) mitogenome (GenBank accession: KJ742384; Zhang et al. Citation2016).
U. erigeronense mitogenome (GenBank accession: MZ695840) is 15,691 bp long, which is longer than S. avenae mitogenome (Zhang et al. Citation2016). It contains 13 protein-coding genes, two ribosomal RNAs, and 22 transfer RNAs, which is a typical configuration of the aphid mitogenomes (Cameron Citation2014; Boore Citation1999). Its nucleotide composition is AT-biased (A + T ratio: 84.2%). Control region is 968 bp long, which is longer than S. avenae mitogenome (430 bp in length). It is the major reason that U. erigeronense mitogenome is longer than S. avenae mitogenome. Most of aphid mitogenomes contain another repeat region between tRNA-Glu and NAD5 (Lee et al. Citation2019; Park, Xi, et al. Citation2019; Park, Kim, et al. Citation2020). The second repeat region of U. erigeronense mitogenome is 245 bp long, which is shorter than that of S. avenae (261 bp).
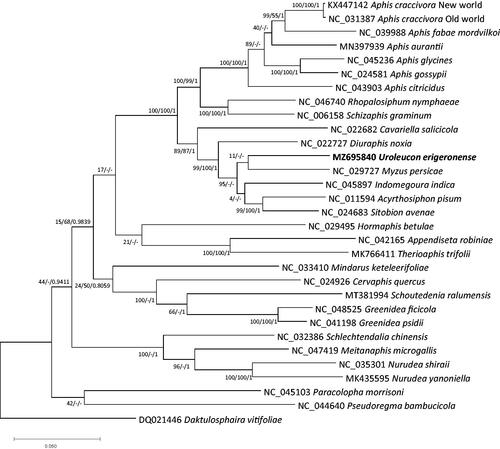
Thirty Aphididae mitogenomes which are representative mitogenome for each species including U. erigeronense mitogenome with one outgroup species, Daktulosphaira vitifoliae (Fitch, 1856) (Hemiptera: Phylloxeridae), were aligned by MAFFT v7.450 (Katoh and Standley Citation2013) for conducting phylogenetic analysis of the Maximum-Likelihood (ML) with 1,000 bootstrap repeats and the Neighbor-Joining (NJ) with 10,000 bootstrap repeats by MEGA X (Kumar et al. Citation2018) and Bayisean inferrence (BI) with GTR model with gamma rates and 1,100,000 generations for Monte Carlo algorithm by MrBayes v3.2.6 (Ronquist et al. Citation2012). Three phylogenetic trees displayed the incongruency of topology of the clade containing to U. erigeronense: the ML tree presented that U. erigeronense was clustered with Myzus persicae (Sulzer, 1776) with very low supportive values (); while NJ and BI trees exhibited that U. erigeronense was clustered with Acrythosiphon pisum (Harris, 1776) and S. avernae mitogenomes with high supportive values, congruent to the molecular phylogeny of nuclear and mitochondrial marker sequences (Moran et al. Citation1999). Moreover, additional clades displaying incongruence among three trees were also found (), indicating that additional studies for clarifying phylogenetic tree of aphid species are required. Taken together, the U. erigeronense mitogenome sequence addresses the further questions to understand phylogenetic relationship and mitogenome characteristics in tribe Macrosiphini in near future.
Figure 1. Maximum-Likelihood, Neighbor-Joining, and Bayesian inference phylogenetic trees of 31 mitochondrial genomes of Aphididae and one outgroup species, D. vitifoliae. Phylogenetic tree was drawn based on Maximum-Likelihood tree. The numbers above branches indicate supportive values of Maximum-Likelihood and Bayesian inference phylogenetic trees, respectively.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The mitochondrial genomes in this study can be accessed via the NCBI GenBank accession number, MZ695840. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA756639, SAMN20893750, and SRR15558516, respectively.
Additional information
Funding
References
- Bae Y, Park J, Lee W. 2020. The complete mitochondrial genome of Aphis gossypii Glover, 1877 (Hemiptera: Aphididae) isolated from Plantago asiatica in Korea. Mitochondrial DNA B Resour. 5(3):2878–2880.
- Blackman RL, Eastop VF. 2006. Aphids on the world’s herbaceous plants and shrubs. West Sussex, U.K: John Wiley & Sons, Ltd. p. 1439.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780. doi:https://doi.org/10.1093/nar/27.8.1767.
- Brumley C, Watson L. 2017. First records of Uroleucon erigeronense (Hemiptera: Aphididae) on Conyza (Asteraceae) from Australia, with descriptions of morphological variation, biological notes and an update for commonly used keys. Austral Entomol. 56(3):339–344.
- Cameron SL. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59:95–117. doi:https://doi.org/10.1146/annurev-ento-011613-162007.
- Choi NJ, Xi H, Park J. 2021. A comparative analyses of the complete mitochondrial genomes of fungal endosymbionts in Sogatella furcifera, white-backed planthoppers. Int J Genomics. 2021:1–20.
- Han T, Park H, Lee J-H, Hong K-J, Kim Y, Park J, Lee W. 2017. The complete mitochondrial genome of the subterranean termite, Reticulitermes kanmonensis Takematsu, 1999 (Isoptera: Rhinotermitidae). Mitochondrial DNA B Resour. 2(2):508–509.
- Jensen AS, Miller GL, Carmichael A. 2010. Host range and biology of Uroleucon (Lambersius) erigeronense (Thomas 1878), and its synonymy with Uroleucon (Lambersius) escalantii (Knowlton) 1928 (Hemiptera: Aphididae). Proc Entomol Soc Washington. 112(2):239–245.
- Jung Y-J, Jo J, Bae Y, Xi H, Seol M-A, Yoo S-H, Park J, Park C. 2021. The complete mitochondrial genome of Myzus persicae (Sulzer, 1776; Hemiptera: Aphididae) isolated in Korea. Mitochondrial DNA B Resour. 6(1):10–12.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Lee H, Park J, Xi H, Lee G-S, Kim I, Park J, Lee W. 2020. The complete mitochondrial genome of Ricania speculum (Walker, 1851) (Hemiptera: Ricaniidae): investigation of intraspecific variations on mitochondrial genome. Mitochondrial DNA Part B. 5(3):3796–3798.
- Lee J, Park J, Lee H, Park J, Lee W. 2019. The complete mitochondrial genome of Paracolopha morrisoni (Baker, 1919) (Hemiptera: Aphididae). Mitochondrial DNA B Resour. 4(2):3037–3039.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079.
- Moran NA, Kaplan ME, Gelsey MJ, Murphy TG, Scholes EA. 1999. Phylogenetics and evolution of the aphid genus Uroleucon based on mitochondrial and nuclear DNA sequences. Systematic Entomology. 24(1):85–93.
- Nafría JMN, Durante MPM, Ortego J, Fernández MVS. 2007. The genus Uroleucon (Hemiptera: Aphididae: Macrosiphini) in Argentina, with descriptions of five new species. Can Entomol. 139(2):154–178.
- Park J, An J-H, Kim Y, Kim D, Yang B-G, Kim T. 2020. Database of National Species List of Korea: the taxonomical systematics platform for managing scientific names of Korean native species. J Species Res. 9(3):233–246.
- Park J, Jung J-K, Koh H, Park J, Seo B-Y. 2019. The complete mitochondrial genome of Laodelphax striatellus (Fallén, 1826) (Hemiptera: Delphacidae) collected in a mid-Western part of Korean peninsula. Mitochondrial DNA Part B. 4(2):2229–2230.
- Park J, Kim Y, Xi H, Park J, Lee W. 2020. The complete mitochondrial genome of Rhopalosiphum nymphaeae (Linnaeus, 1761) (Hemiptera: Aphididae). Mitochondrial DNA Part B. 5(2):1613–1615.
- Park J, Lee J, Lee W. 2021. The complete mitochondrial genome of Aphis gossypii Glover, 1877 (Hemiptera: Aphididae) isolated from Leonurus japonicus in Korea. Mitochondrial DNA B Resour. 6(1):62–65.
- Park J, Park J. 2021. Complete mitochondrial genome of the jet ant Lasius spathepus Wheeler, WM, 1910 (Formicidae; Hymenoptera). Mitochondrial DNA B Resour. 6(2):505–507.
- Park J, Park J, Kim J-H, Cho JR, Kim Y, Seo BY. 2019. The complete mitochondrial genome of Micromus angulatus (Stephens, 1836) (Neuroptera: Hemerobiidae). Mitochondrial DNA Part B. 4(1):1467–1469.
- Park J, Xi H, Kim Y, Kim M. 2021. Complete genome sequence of Lentilactobacillus parabuchneri Strain KEM. Microbiol Resour Announc. 10(20):e01208–01220.
- Park J, Xi H, Kim Y, Park J, Lee W. 2019. The complete mitochondrial genome of Aphis gossypii Glover, 1877 (Hemiptera: Aphididae) collected in Korean peninsula. Mitochondrial DNA B Resour. 4(2):3007–3009.
- Park J, Xi H, Park J. 2020. The complete mitochondrial genome of Rotunda rotundapex (Miyata & Kishida, 1990) (Lepidoptera: Bombycidae). Mitochondrial DNA B Resour. 5(1):355–357.
- Park J, Xi H, Park J. 2021. Complete genome sequence of a Blochmannia endosymbiont of Colobopsis nipponica. Microbiol Resour Announc. 10(17):e01195–01120.
- Park J, Xi H, Park J, Lee W. 2020. The complete mitochondrial genome of fungal endosymbiont, Ophiocordycipitaceae sp., isolated from Ricania speculum (Hemiptera: Ricaniidae). Mitochondrial DNA Part B. 5(2):1888–1889.
- Park J, Xi H, Park J, Nam SJ, Lee Y-D. 2020. Complete genome sequence of the Blochmannia endosymbiont of Camponotus nipponensis. Microbiol Resour Announc. 9(29):e00703–00720.
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829.
- Zhang B, Zheng J, Liang L, Fuller S, Ma C-S. 2016. The complete mitochondrial genome of Sitobion avenae (Hemiptera: Aphididae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(2):945–946.
- Zhao QY, Wang Y, Kong YM, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinf. 12(S14):S2.
