Abstract
Gymnocorymbus ternetzi belongs to the genus Gymnocorymbus in the family Characidae, and is mainly distributed in southern Brazil. Herein, we report the complete mitogenome of G. ternetzi using Illumina sequencing data. The mitogenome is 17,999 bp in length and contains 13 protein-coding genes, 22 transfer RNA genes, and 2 ribosomal RNA genes. Phylogenetic analysis of G. ternetzi and 18 related species within Characidae indicates that G. ternetzi clusters within the family Characidae. The data provide useful genetic information for future studies on the taxonomy, phylogeny, and evolution of Characidae species.
Characidae is among the most diverse families of Characiformes and one of the largest clades of fish globally, with a complex taxonomic background (Benine et al. Citation2015). The black widow tetra Gymnocorymbus ternetzi (Boulenger, 1895), belonging to the genus Gymnocorymbus within the family Characidae, is a peaceful shoaling tropical freshwater fish distributed in the rivers of southern Brazil (Priestley et al., Citation2006). It is an omnivorous fish that feeds on plants, insects, and crustaceans (Polaz et al. Citation2017). Because of its attractive appearance, undemanding maintenance, and ease of breeding, G. ternetzi is a popular freshwater ornamental fish (Frankel Citation2004; Uma and Chandran Citation2008). The evolutionary genetics of G. ternetzi are unknown. In this study, we report the complete mitochondrial genome of this species. The data will be a valuable resource for further studies on evolutionary genetics, species delimitation, and phylogenetic studies on Gymnocorymbus.
Samples were collected from Pudong New Area, Shanghai City, China (31°19′06.89″N, 121°67′90.75″E). The voucher specimen was deposited at the Lishui University (No. LSU-ZJ2021-03-01–LSU-ZJ2021-03-10 by Ziming Liu, [email protected]). Genomic DNA was extracted using a TIANamp Genomic DNA Kit (TIANGEN, Beijing, China). The sequencing library was produced using the Illumina Truseq™ DNA Sample Preparation Kit (Illumina, San Diego, CA, USA) according to the manufacturer's recommendations. The prepared library was loaded on an Illumina Novaseq 6000 platform for paired-end 2 × 150 bp sequencing at Novogene (Beijing, China). Raw data were used to assemble the complete mitogenome genome using the GetOrganelle pipeline (Jin et al. Citation2020). Genome annotation was performed using Mitoz annotation module (Meng et al. Citation2019). The annotated genome sequence was deposited in GenBank under accession number MZ363625.
The circular mitogenome of G. ternetzi is 17,999 bp in length, with 29.92% A, 28.41% T, 15.37% G, and 26.30% C. The greater A + T content (58.33%) than the G + C content (41.67%) indicated a slight A + T bias in G. ternetzi. Further, 37 genes were predicted, including 13 protein-coding genes, 22 transfer RNAs, and 2 ribosomal RNA genes.
Phylogenetic analysis was performed using complete mitogenomes from 23 species of Paralichthys olivaceus and Cynoglossus semilaevis serving as outgroup taxa. The genomes were aligned with MAFFT v7.388 using default settings (Katoh and Standley Citation2013). Phylogenetic analysis was conducted based on maximum likelihood (ML) analyses implemented in IQ-TREE v2.1.2 with the GTR + F+R2 nucleotide substitution model selected by ModelFinder (Kalyaanamoorthy et al. Citation2017; Minh et al. Citation2020). Support for the inferred ML tree was inferred by bootstrapping with 1,000 replicates. The analysis showed that G. ternetzi was in a clade with Gephyrocharax atracaudatus ().
Figure 1. Maximum likelihood (ML) tree based on 19 mitogenome sequences of representative fish in Characidae as the ingroup and Paralichthys olivaceus and Cynoglossus semilaevis as the outgroup. Numbers on the nodes are bootstrap values based on 1,000 replicates. The G. ternetzi genome is marked in bold and red font.
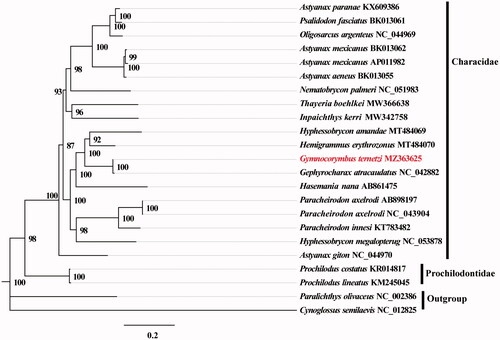
This study provides important sequence information for species identification and phylogenetic relationships in the Characidae species.
Disclosure statement
No potential conflict of interest is reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank (https://www.ncbi.nlm.nih.gov/) under accession no MZ363625. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA725085, SAMN18869625, and SRR14322678, respectively.
Additional information
Funding
References
- Benine RC, Melo BF, Castro RMC, Oliveira C. 2015. Taxonomic revision and molecular phylogeny of Gymnocorymbus Eigenmann, 1908 (Teleostei, Characiformes, Characidae) ). Zootaxa. 3956(1):1–28.
- Frankel JS. 2004. Inheritance of trunk banding in the tetra (Gymnocorymbus ternetzi Characidae). J Hered. 95(3):262–264.
- Jin JJ, Yu WB, Yang JB, Song Y, Depamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):1–31.
- Kalyaanamoorthy S, Minh BQ, Wong TK, Von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Meng GL, Li YY, Yang CT, Liu SL. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids RES. 47(11):63.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534.
- Polaz CNM, Ferreira FC, Petrere JM. 2017. The protected areas system in Brazil as a baseline condition for wetlands management and fish conservancy: the example of the Pantanal National Park. Neotrop Ichthyol. 15(3):e170041.
- Priestley SM, Stevenson AE, Alexander LG. 2006. Growth rate and body condition in relation to group size in Black Widow Tetras (Gymnocorymbus ternetzi) and Common Goldfish (Carassius auratus). J Nutr. 136(7 Suppl):2078S–2080S. doi:https://doi.org/10.1093/jn/136.7.2078S. 16772504
- Uma B, Chandran MR. 2008. Induction of triploidy in Gymnocorymbus Ternetzi (Boulenger). Res J Fish Hydrobiol. 3(2):41–47.
