Abstract
Ulmus lanceifolia, which plays an important role in ecology and economy, is one of the main evergreen broad leaved arbor species in southern China. In this study, we assembled its complete chloroplast genome. The total genome size of U. lanceifolia was 158,652bp, containing a large single-copy region of 87,119bp, a small single-copy region of 18,697bp, and a pair of inverted repeat regions of 26,418bp. The overall GC content of U. lanceifolia chloroplast genome is 35.63%. We obtained the 132 genes chloroplast genome, including 86 proteincoding genes, 8 rRNA genes, and 37 tRNA genes. The phylogenetic analysis suggests that U. lanceifolia is a monophyletic taxa and a transitional species between the 2 groups which were combined by Ulmus americana, U. elongate and Zelkova serrate, Z. schneideriana respectively in Ulmaceae.
Ulmus lanceifolia (Roxburgh ex Wallich Citation1831) is one of the main evergreen broad leaved arbor species in southern China, with fairly good material toughness. It is around 30 meters tall, with a diameter of 40–80 centimeters at breast hight. Bark yellowish gray to chestnut brown, exfoliating in irregular flakes (Fu et al. Citation2003). The timber of U. lanceifolia is heavy and hard, not only is a good material for furniture, indoor decoration and plywood, but also suitable for vehicles, ships, farm tools, sporting goods and musical instruments (Xiao and Mu Citation1998). In addition, U. lanceifolia also has great potential in medicinal application. But to date, there is still no complete chloroplast (cp) genome is characterized for U. lanceifolia. The object of this work was to explore the intrinsic distinction in an effort to vindicate its taxonomic status in Ulmaceae. Here we present the cp genome of U. lanceifolia (GeneBank accession number: MZ159247) as an initial approach toward developing genomic tools to better understand U. lanceifolia and its close relatives.
The fresh leaves of U. lanceifolia were sampled from Xishuangbanna Tropical Botanical Garden (**latitude**101.262951, **longitude**21.931604), Yunnan, China. A specimen was deposited at the herbarium of Nanjing Forestry University (contact person: [email protected]) under the voucher number NF2021039. The whole genome sequencing was conducted using the Illumina NovaSeq platform by Nanjing Genepioneer Biotechnology Inc. (Nanjing, China). The raw reading was filtered by software fastp version 0.20.0, and the clean reading was used to assemble the chloroplast genome by SPAdes v3.10.1 (Bankevich et al. Citation2012). Finally, the gene structure was annotated using CpGAVAS2(Liu et al. Citation2012). The 20 complete chloroplast sequences were aligned by the MAFFT v7.475 (Katoh et al. Citation2019). Then we generated a maximum-likelihood (ML) tree through IQ TREE (Nguyen et al. Citation2015) with 1000 bootstrap alignments.
The complete chloroplast genome of U. lanceifolia was determined to be 158,652bp in length and contained 2 inverted repeat (IRa and IRb) regions of 26,418bp. The repeat regions divided the genome into 2 single-copy regions, SSC and LSC with 18,697bp and 87,119bp. A total of 132 genes are encoded, including 86 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. Most of genes occurred in a single copy; however, 7 protein-coding genes (ndhB, rpl2, rpl23, rps12, rps7, ycf15 and ycf2), 7 tRNA genes (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG and trnV-GAC) and 4 distinct rRNA gene (23S, 16S, 5S and 4.5S) are duplicated. A total of 18 protein-coding genes (atpF, ndhA, ndhB, petB, petD, rpl16, rpl2, rpoC1, rps12, rps16, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, trnV-UAC) contained 1 intron while the other 2 genes (clpP, ycf3) had 2 intron each. The overall GC content of U. lanceifolia genome was 35.63%, and the corresponding values in LSC, SSC and IR regions were 33.14%, 28.34%, and 42.33%, respectively.
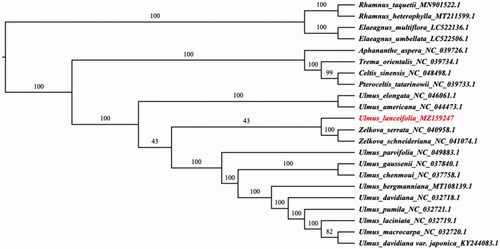
The ML analysis reveals that U. lanceifolia is a monophyletic taxa and a transitional species between the 2 groups which were combined by Ulmus americana, U. elongate and Zelkova serrate, Z. schneideriana respectively in Ulmaceae ().
Disclosure statement
No potential conflict of interest was reported by the authors.
The data availability statement
The data that support the findings of this study are openly available in the Genbank database at https://www.ncbi.nlm.nih.gov/, under accession number [MZ159247]. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA753672, SRR15412756, and SAMN20695145 respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Fu L-K, Xin Y-Q, Alan W. 2003. Ulmaceae. In: Wu Z-Y, Raven PH & Hong D-Y, editors. Flora of China, vols. 5 (Ulmaceae). Beijing; St. Louis: Science Press; Missouri Botanical Garden. p. 1–10.
- Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4):1160–1166.
- Liu C, Shi L-C, Zhu Y-J, Chen H-M, Zhang J-H, Lin X-H, Guan X-J. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Roxburgh ex Wallich. 1831. Plantae Asiaticae Rariores: or, Descriptions and figures of a select number of unpublished East Indian plants. Roxburgh ex W. 2(8):86.
- Xiao S-Q, Mu Q-Y. 1998. Wood structure of evergreen elm (Ulmus lanceaefolia) and classification of the commercial timber in Chinese elm. J Sichuan Agri Univ. 16(01):95–98.