Abstract
Fopius arisanus (Sonan, 1932), an important egg parasitoid of several notorious tephritid pests, plays a key role in biological control programs. In the present study, the whole mitochondrial genome of F. arisanus was sequenced and characterized. The mitogenome of F. arisanus is 16,425 bp in length with 14.94% GC content, and contains 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), and two ribosomal RNA genes (rRNAs). The phylogenetic trees demonstrated that F. arisanus is sister group to Psyttalia concolor, P. humilis, P. lounsburyi and Diachasmimorpha longicaudata.
Fopius arisanus (Sonan, Citation1932) (Hymenoptera: Braconidae) parasitizes more than 40 species of tephritid pest (Cai et al. Citation2017). Owing to its unique characteristics for parasitizing eggs, F. arisanus has become a key component of biological control programs that aim to suppress tephtirid pest populations and therefore reduce economic loss (Vargas et al. Citation2001). However, to date, there are few studies detailing its genome information. Hence, in this study, we determined the complete mitochondrial genome of F. arisanus and analyzed the evolutionary relationship between F. arisanus and other braconid wasps.
The samples were obtained from Fujian Agriculture and Forestry University (26.084220°N, 119.231164°E), Fuzhou City, Fujian Province, China. The voucher specimens (20190808FA) were deposited at the Fujian Agriculture and Forestry University (URL: http://zbxy.fafu.edu.cn; contact: Qinge Ji, [email protected]). Total DNA was extracted using CTAB extraction method (Vanzyme, Nanjing, China) and a 400-bp insert library was constructed. An Illumina Novaseq 6000 platform in 150 bp paired-end read mode was used to sequence the constructed library. Filtering of raw data was performed in fastp v.0.20.0 (Chen et al. Citation2018) resulting in 21,648,152 clean reads, which were then assembled by SPAdes v.3.9.0 software (Bankevich et al. 2012). Annotation of the assembled sequence was performed using MITOS web server (Bernt et al. Citation2013).
The complete mitochondrial genome of F. arisanus is 16,425 bp in length, containing 13 protein-coding genes (PCGs), two ribosomal RNA genes (rRNAs), 22 transfer RNA genes (tRNAs), and a non-coding region (control region). The mitogenome comprises 39.37% A, 9.14% G, 5.8% C, and 45.68% T, with a significant A + T (85.06%) bias. For the PCGs, six genes (cox2, atp8, nad2, nad4l, nad5, nad6) had a start codon of ATT, four (cox1, cox3, atp6, cob) had ATG, and three had ATA (nad1, nad3, nad4). All PCGs contained the stop codon TAA, except for nad3 which had TAG.
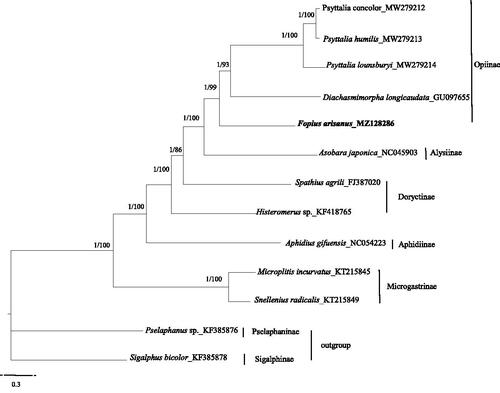
We analyzed the nucleotide sequences of PCGs using the Maximum-Likelihood (ML) and Bayesian Inference (BI) approaches to understand the phylogenetic relationship of F. arisanus with 10 other species and to estimate the phylogenetic position of F. arisanus. Two species respectively from Sigalphinae and Pselaphaninae were used as outgroups (Lyu et al. Citation2020). Phylogenetic analyses were performed with Bayesian inference in MrBayes 3.2.3 (Ronquist et al. Citation2012) and maximum likelihood in RAxML 8.2.10 (Stamatakis Citation2014). The phylogenetic trees showed that F. arisanus clustered with Psyttalia concolor, P. humilis, P. lounsburyi and Diachasmimorpha longicaudata as a separated clade. To date, studies that relate to the genome analysis of F. arisanus are limited, and therefore we hope that our data provides valuable information for further studies .
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The complete mitochondrial genome sequence of F. arisanus is deposited in the GenBank database under the accession number MZ128286. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA734964, SRS9131624, and SAMN19551218, respectively The Web link is https://www.ncbi.nlm.nih.gov/
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler P F. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2): 313–319.
- Cai PM, Gu XH, Yao MY, Zhang HH, Huang J, Idress A, Ji QE, Chen JH, Yang JQ. 2017. The optimal age and radiation dose for Bactrocera dorsalis (Hendel) (Diptera: Tephritidae) eggs as hosts for mass-reared Fopius arisanus (Sonan) (Hymenoptera: Braconidae). Biol. Control. 108:89–97.
- Chen SF, Zhou YQ, Chen Y, Gu J. 2018. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34(17):i884–i890.
- Lyu BQ, Meng R, Cai B, Su H. 2020. Complete mitochondrial genome of a parasitic wasp Microplitis pallidipes (Hymenoptera: Braconidae: Microgastrinae). Mitochondrial DNA B Resour. 5(1):871–872.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Sonan J. 1932. Notes on some Braconidae and Ichneumonidae from Formosa, with descriptions of 18 new species. Trans Nat Hist Soc Formose. 22:69–82.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Vargas RI, Peck SL, McQuate GT, Jackson G, Stark JD, Armstrong JW. 2001. Potential for areawide integrated management of Mediterranean fruit fly (Diptera: Tephritidae) with a braconid parasitoid and a novel bait spray. J Econ Entomol. 94(4):817–825.