Abstract
Here, we used RNA-seq reads to assemble the complete mitochondrial genomes of the spring field cricket, Gryllus veletis, and the variable field cricket, Gryllus lineaticeps. The mitochondrial genomes of G. veletis (15,686 bp, MW322713) and G. lineaticeps (15,607 bp, MW315773) each contain the expected 13 protein-coding genes, two ribosomal RNA genes, 22 transfer RNA genes, and a large control (D-loop) region. The arrangements of these features were similar for both species and consistent with other closely related Orthoptera. A phylogenetic analysis of the mitochondrial genome sequences revealed that G. veletis and G. lineaticeps cluster with the other Gryllus species and all reside in a clade with the Gryllidae.
Field crickets (Gryllus spp.; Orthoptera: Gryllidae) have a Holarctic distribution and are often subjects of evolutionary, behavioral, physiological, and acoustic studies (e.g. Adamo and Baker Citation2011; Blankers et al. Citation2017; Rodríguez-Muñoz et al. Citation2008). The fall field cricket, Gryllus veletis Alexander and Bigelow Citation1960, is an emerging model for insect freeze tolerance (Toxopeus et al. Citation2018b) that is broadly distributed in North America, whereas the variable field cricket, Gryllus lineaticeps Stål, 1861, featured in behavior (e.g. Wagner Citation1996) and physiology (e.g. Sun et al. Citation2020) studies, is restricted to California and Southern Oregon (Alexander and Bigelow 1960; Weissman and Gray Citation2019). We collected a representative G. veletis sample from a lab-reared colony at the University of Western Ontario, which was derived from samples originally collected from the University of Lethbridge campus in Lethbridge, Alberta, Canada (49.68° N, 112.86° W). The representative G. lineaticeps sample was collected from a colony maintained from field-collected individuals from Sedgwick Reserve, Santa Ynez, California, USA (34°42′N 120°1′W).
We assembled the mitochondrial genome sequences using RNA-seq reads as described by Tian and Smith (Citation2016). Briefly, contiguous RNA sequences aligning to mitochondrial DNA sequences were mined from a de novo transcriptome assembly of G. veletis (NCBI Bioproject accession: PRJNA479659; Toxopeus et al. Citation2018a) using a nucleotide BLAST (blastn). The mitochondrial RNA-derived contigs were then mapped to the mitochondrial genome of Gryllus bimaculatus (Genbank accession: MK204367; Wang et al. Citation2019) using Geneious Prime v2020.2 (https://www.geneious.com). We mapped the G. lineaticeps reads onto the G. bimaculatus mitochondrial genome because a transcriptomic assembly has not yet been published. For both species, we resolved regions that did not map to the reference mitochondrial genome by remapping raw reads to the existing assembled contigs and relied on the overhang of newly mapped reads to extend the sequences until the sequence would no longer extend. We repeated this process iteratively to yield a complete mitochondrial sequence. We annotated the complete mitochondrial genomes using Geneious and tRNAscan-SE version 2.0 (Lowe and Chan Citation2016). The voucher specimens for G. lineaticeps (CNC1150967, CNC1150968, CNC1150969) and G. veletis (CNC1150970, CNC1150971) were deposited at the Canadian National Collection of Insects, Arachnids, and Nematodes (https://www.agr.gc.ca/eng/scientific-collaboration-and-research-in-agriculture/agriculture-and-agri-food-research-centres-and-collections/canadian-national-collection-of-insects-arachnids-and-nematodes-cnc/; Owen Lonsdale; [email protected]).
The lengths of the mtDNA sequences from G. veletis and G. lineaticeps are 15,686 and 15,607 bp, respectively. These lengths are shorter than G. bimaculatus (16,301 bp), but comparable to related crickets in Gryllidae (e.g. Teleogryllus emma; 15,697 bp). Each mitochondrial genome contains the expected set of 37 genes consisting of 13 protein-coding genes (ND1-6, ND4L, COX1-3, ATP8, ATP6, and CYTB), 12S and 16S rRNAs, 22 tRNAs and a putative A + T rich (D-loop) control region. The D-loop measured 933 and 816 bp in G. veletis and G. lineaticeps, respectively. The overall base composition for G. veletis is A: 40.1%, C: 16.9%, G: 9.4%, and T: 33.5% and for G. lineaticeps is A: 39.9%, C: 17.8%, G: 9.7%, and T: 32.7%. These distributions are similar to all of the species used in our comparative analysis.
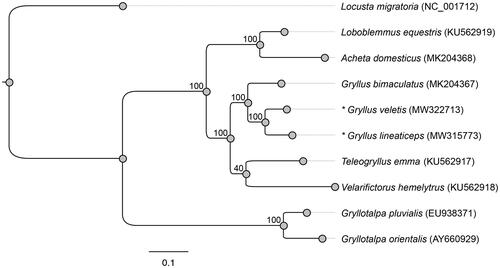
We compared the phylogenetic relationships of the two newly sequenced mitochondrial genomes with those of eight other Orthoptera presented by Wang et al. (Citation2019) (). We constructed the phylogenetic relationships among these species using the maximum likelihood method in Geneious. We confirmed that G. veletis and G. lineaticeps are clustered with Gryllus bimaculatus (Wang et al. Citation2019) and rooted with other Gryllidae species (Loxoblemmus equestris, Acheta domesticus, Velarifictorus hemelytrus, and Teleogryllus emma). Consistent with the nuclear DNA phylogeny published by Gray et al. (Citation2020), Gryllus crickets appear to be more closely related to Teleogryllus than to Acheta.
Figure 1. Phylogenetic positions of Gryllus veletis and Gryllus lineaticeps based on the complete mitochondrial genomes of seven other Orthoptera constructed using maximum likelihood. The numerical values indicate bootstrap support for each node (100 permutations). Each Latin name is followed by the respective mitochondrial genome GenBank accession number. The focal species of this study are denoted with an asterisk.
Acknowledgements
We would like to thank Jared Deyarmin and Zachary Stahlschmidt for providing access to the G. lineaticeps RNA-seq data. We also thank Zachary Stahlschmidt and Alyssa Stevens for preparing the voucher specimens for G. lineaticeps and G. veletis, respectively, and four anonymous reviewers for feedback that improved the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The complete mtRNA sequences for G. veletis (MW322713) and G. lineaticeps (MW315773) are available through GenBank of NCBI at [https://www.ncbi.nlm.nih.gov]. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA479659, SRP151981, and SAMN09598196-SAMN09598198, respectively.
Additional information
Funding
References
- Adamo SA, Baker JL. 2011. Conserved features of chronic stress across phyla: the effects of long-term stress on behavior and the concentration of the neurohormone octopamine in the cricket, Gryllus texensis. Horm Behav. 60:478–483.
- Alexander RD, Bigelow RS. 1960. Allochronic speciation in field crickets, and a new species. Acheta veletis. Evolution. 14:334–346.
- Blankers T, Gray DA, Hennig RM. 2017. Multivariate phenotypic evolution: divergent acoustic signals and sexual selection in Gryllus field crickets. Evol Biol. 44:43–55.
- Gray DA, Weissman DB, Cole JA, Lemmon EM. 2020. Multilocus phylogeny of Gryllus field crickets (Orthoptera: Gryllidae: Gryllinae) utilizing anchored hybrid enrichment. Zootaxa. 4750:328–348.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44:W54–W57.
- Rodríguez-Muñoz R, Bretman A, Hadfield JD, Tregenza T. 2008. Sexual selection in the cricket Gryllus bimaculatus: no good genes? Genetica. 132:287–294.
- Sun BJ, Huebner C, Treidel LA, Clark RM, Roberts KT, Kenagy GJ, Williams CM, Overgaard J. 2020. Nocturnal dispersal flight of crickets: behavioural and physiological responses to cool environmental temperatures. Funct Ecol. 34:1907–1920.
- Tian Y, Smith DR. 2016. Recovering complete mitochondrial genome sequences from RNA-Seq: a case study of Polytomella non-photosynthetic green algae. Mol Phylogenet Evol. 98:57–62.
- Toxopeus J, Des Marteaux LE, Sinclair BJ. 2018a. How crickets become freeze tolerant: The transcriptomic underpinnings of acclimation in Gryllus veletis. Comp Biochem Physiol D . 29:55–66.
- Toxopeus J, McKinnon AH, Štětina T, Turnbull KF, Sinclair BJ. 2018b. Laboratory acclimation to autumn-like conditions induces freeze tolerance in the spring field cricket Gryllus veletis (Orthoptera: Gryllidae). J Insect Physiol. 113:9–16.
- Wagner WE. Jr, 1996. Convergent song preferences between female field crickets and acoustically orienting parasitoid flies. Behav Ecol. 7:279–285.
- Wang C, Li QY, Xu CL, Liu G. 2019. The complete mitochondrial genome of two-spotted cricket Gryllus bimaculatus (Grylloidea: Gryllidae). Mitochondr DNA B. 4:799–800.
- Weissman DB, Gray DA. 2019. Crickets of the genus Gryllus in the United States (Orthoptera: Gryllidae: Gryllinae). Zootaxa. 4705:1–277.
