Abstract
Primulina tenuituba is a species in the Gesneriaceae family that is widely distributed in China. It is a karst-dwelling species with an enormous tolerance for extreme drought and high temperatures. The species is also used in traditional Chinese medicine. In this study, the complete chloroplast genome of P. tenuituba was assembled and characterized for the first time. The complete chloroplast genome exhibited a typical quadripartite cycle of 153,236 bp in length, including a pair of inverted repeats (IRs) of 25,494 bp, which were separated by a large single-copy (LSC) region of 84,364 bp and a small single copy (SSC) region of 17,884 bp. The GC content was 37.6%. The complete chloroplast genome of P. tenuituba contains 114 genes, including 80 protein-coding genes, 30 tRNAs genes, and four rRNAs. The phylogenetic analysis showed that P. tenuituba is closely related to P. eburnea. The newly reported chloroplast genome of P. tenuituba would provide valuable data for further studies on its evolution and adaptation mechanism.
Primulina tenuituba (W.T. Wang) Yin Z. Wang (Gesneriaceae), a karst-dwelling species, is widely distributed in Guizhou, Hunan, Sichuan, and Hubei Provinces in China, and appears to have a strong tolerance for extreme drought and high temperatures. Its rhizome is also used in traditional Chinese medicine in treating rheumatism (Editorial Committee of the Flora of China Citation1990; Li and Wang Citation2004). However, its phylogenetic relationship in the Gesneriaceae family is unclear due to the absence of genome (plastome) information. After our research, we can now report the complete chloroplast genome of P. tenuituba to clarify its phylogenetic placement in Gesneriaceae. The plants were collected from Guiyang, Guizhou, China (26°35′4.43″ N, 106°43′35.20″ E), and a voucher specimen (WF200808-12) was deposited at the herbarium of Guizhou Normal University (GNUB, https://sjxy.gznu.edu.cn/, and emailed to contact person Tao Peng, [email protected]). Total genomic DNA was extracted from the fresh leaves of P. tenuituba using a modified cetyltrimethylammonium bromide (CTAB) method (Doyle and Doyle Citation1987) and sent to Majorbio Company (http://www.majorbio.com/, China) for next-generation sequencing. Short-insert (350 bp) paired-end read libraries preparation and 2 × 150 bp sequencing were performed on an Illumina (HiSeq4000) genome analyzer platform. A total of 13,101,144 raw reads were obtained, and 98.27% of which were declared as clean reads. The clean reads were mapped to P. ophiopogoides (D. Fang & W.T. Wang) Yin Z. Wang (MF227819, complete chloroplast genome reference) in Geneious Primer (Kearse et al. Citation2012) to exclude nuclear and mitochondrial reads. Then de novo was performed for assembling construction. Contigs were concatenated using the Repeat Finder option implemented in Geneious Primer until only one contig (including SSC, IR, and LSC) was built. The IR region was detected by the Repeat Finder option in Geneious Primer and was reverse copied to obtain the complete plastid genome. The annotation approach was performed using CPGAVAS2 and PGA (Qu et al. Citation2019; Shi et al. Citation2019).
The complete chloroplast genome of P. tenuituba was 153,236 bp in length (GenBank-MW245830), containing one large single-copy (LSC) region of 84,364 bp and one small single-copy (SSC) region of 17,884 bp, which are separated by two inverted repeat (IR) regions of 25,494 bp. The GC content was 37.6%. A total of 114 unique genes were annotated in the complete chloroplast genome, including 80 protein-coding genes, four ribosomal RNA genes, and 30 transfer RNA genes.
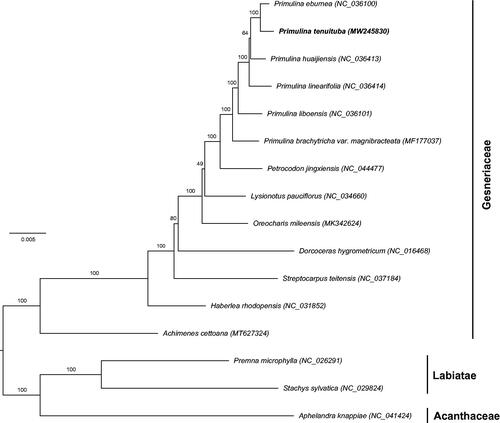
To investigate the phylogenetic relationships with the closely related species, the maximum likelihood (ML) phylogenetic tree was constructed by RAxML (Stamatakis Citation2006) based on 13 complete chloroplast genomes in Gesneriaceae (). Aphelandra Knappiae Wassh. representing Acanthaceae and Premna microphylla Turcz. and Stachys sylvatica L. representing Labiatae were employed as outgroups. The result showed that Gesneriaceae is monophyletic. Primulina tenuituba fell into a monophyletic group Primulina and was closely related to P. eburnea (Hance) Yin Z. Wang. This relationship was congruent with their morphological similarity, such as the narrowly long leaves. The newly reported complete chloroplast genome of P. tenuituba would provide valuable data for further studies on its evolution and adaptation mechanism.
Acknowledgments
We want to thank Michael LoFurno, Adjunct Professor, Temple University, and Stephen Maciejewski, the Gesneriad Society, Philadelphia, the USA, for their editorial assistance.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data supporting this study’s findings are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW245830. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA682283, SRR13188905, and SAMN16986174, respectively.
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Editorial Committee of the Flora of China. 1990. Flora of China. Vol. 69. Beijing (China): Science Press; p. 392.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Li ZY, Wang YZ. 2004. Plants in the family Gesneriaceae in China. Zhengzhou (China): Henan Science and Technology Press; p. 249.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Meth. 15:50.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690.