Abstract
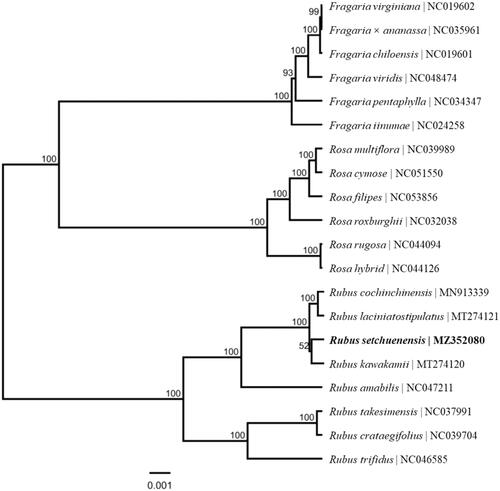
Rubus setchuenensis Bureau et Franch. is important in phylogeny and evolution amongst genus Rubus L. (Rosaceae) plants. The chloroplast genome of R. setchuenensis reported in this study is 156,231 bp in size, with an average GC content of 37.19%. The complete chloroplast genome has a typical quadripartite structure, including a large single copy (LSC) region (85,829 bp) and a small single copy (SSC) region (18,860 bp), which are separated by a pair of inverted repeats (IRs, 25,771 bp). This plastome contains 129 different genes, including 85 protein-coding genes, 36 tRNA genes, and 8 rRNA genes. The phylogenetic analysis of 20 chloroplast genomes from genera Fragaria, Rosa and Rubus of the family Rosaceae suggested that R. setchuenensis clustered into one clade with the other three species of section Malachobatus Focke, and then grouped with four species of section Idaeobatus Focke, while species from Fragaria and Rosa were classified into a group, separately.
Rubus setchuenensis Bureau et Franch. belongs to genus Rubus L., section Malachobatus Focke, subsection Moluccani (Focke) Yu et Lu in the family Rosaceae, it is widely distributed in temperate zone, subtropical and tropical zone in China, such as Hubei, Hunan, Guangxi, Sichuan, Yunnan and Guizhou province, with elevation between 500 and 3000 m (Robertson Citation1974; Yu and Lu Citation1985; Thompson Citation1995; Lu and Boufford Citation2003). Its fresh black fruits can be used for food, and the roots can be used for medicine and tannin extraction. Its bark can be used as a raw material for papermaking, and the seeds can be extracted oil. The most prominent trait is that R. setchuenensis is unarmed with no prickles on the stems and twigs of deciduous shrubs. Therefore, it can be used as an important germplasm resource for hybridization to breed thornless blackberry varieties. However, it is extremely difficult to delimit species of genus Rubus because of complex morphological variations (Alice and Campbell Citation1999), inter- and intraspecific hybridization, polyploidization and apomixis. Frequently, R. setchuenensis was confused with some others related kinship species in the section Malachobatus Focke, subsection Moluccani (Focke) Yu et Lu. The chloroplast genome data will help the development of DNA barcode for Rubus L., but the chloroplast genome of R. setchuenensis has not been published. Therefore, we reported the complete sequence of chloroplast genome of R. setchuenensis here to help construct the phylogeny and evolution of Rubus L.
The fresh leaves of R. setchuenensis were collected from Cuihua town, Daguan County, Zhaotong City, Yunnan Province of China (geospatial coordinates: 27°45′14″N, 103°53′51″E; altitude: 1123 m). The sheets of vouchered specimen, Zhu-20201014R03, had been stored at the herbarium HHP-YAU (Herbarium of Horticultural plants, College of Landscape and Horticulture, Yunnan Agricultural University). Total genomic DNA was extracted from fresh leaves by using DNA Plantzol Reagent (Invitrogen, Carlsbad, CA, USA) to construct chloroplast DNA libraries. The extracted DNA was sequenced by Illumina HiSeq Sequencing System (Illumina, San Diego, CA) and shotgun library was constructed. About 2.29 Gb pair-end (150 bp) raw reads were obtained and the low-quality sequences were filtered by using CLC Genomics Workbench v8.0 (CLC Bio, Aarhus, Denmark) to get high-quality clean reads. NOVOPlasty software (Dierckxsens et al. Citation2017) was used to align and assemble cp genome with R. niveus Thunb (MT576936) served as the reference. The complete chloroplast genome of R. setchuenensis was automatically annotated by using CpGAVAS (Liu et al. Citation2012) and then adjusted and confirmed with Geneious 9.1 (Kearse et al. Citation2012). The complete chloroplast genome had been submitted to the GenBank under the accession number of MZ352080. To ascertain the phylogenetic status of R. setchuenensis, the complete chloroplast genome sequences of 20 species from three genus of family Rosaceae, Fragaria, Rosa and Rubus, were aligned by using MAFFT (Katoh and Standley Citation2013), and a maximum likelihood (ML) tree was reconstructed with RAxML8 (Stamatakis Citation2014).
The chloroplast genome of R. setchuenensis is 156,231 bp in size, with an average GC content of 37.19%, exhibiting a typical quadripartite structure comprising a large single-copy (LSC) region of 85,829 bp and a small single-copy (SSC) region of 18,860 bp separated by a pair of identical inverted repeat regions (IRs) of 25,771 bp each. The chloroplast genome contains 129 genes (112 unique), including 85 protein-coding genes (79 unique), 36 tRNA genes (29 unique), and 8 rRNA genes (4 unique).
The ML phylogenetic tree showed that R. setchuenensis was mostly related to R. kawakamii, it clustered into one clade with the other three species of section Malachobatus Focke, and then grouped with four species of section Idaeobatus Focke, and six species of genera Fragaria and Rosa from Rosaceae formed a monophyletic group independently (). This research lays the foundation for further understanding of the chloroplast genome information of Rubus plants, and provides important information for the development and utilization of peculiar plant resources.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The complete chloroplast genome generated for this study has been deposited in GenBank with accession number MZ352080, which is openly available in GenBank of NCBI at website (https://www.ncbi.nlm.nih.gov/). All high-throughput sequencing data files are available from the GenBank Sequence Read Archive (SRA) with accession number: SRR14757449. The associated BioProject and Bio-Sample numbers are PRJNA735803 and SAMN19602743 respectively.
Additional information
Funding
References
- Alice L, Campbell C. 1999. Phylogeny of Rubus (rosaceae) based on nuclear ribosomal DNA internal transcribed spacer region sequences. Am J Bot. 86(1):81–97.
- Dierckxsens N, Mardulyn P, Smits G. 2017. Novoplasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Liu C, Shi LC, Zhu YJ, Chen HM, Zhang JH, Lin XH, Guan XJ. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715.
- Lu LD, Boufford DE. 2003. Rubus Linnaeus. In: Wu ZY, Raven PH, editors. Flora of China 9. St. Louis: Science Press, Beijing & Missouri Botanical Garden Press; p. 195–285.
- Robertson K. 1974. The genera of Rosaceae in the southeastern United States. J Arnold Arboretum. 55:352–360.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Thompson M. 1995. Chromosome numbers of Rubus species at the National Clonal Germplasm Repository. HortSci. 30(7):1447–1452.
- Yu TT, Lu LT. 1985. Rubus Linnaeus. In: Yu TT, editor. Flora Reipublicae Popularis Sinicae 37. Beijing (China): Science Press; p. 10–218.