Abstract
Baikal sculpins are the most species-rich and ecologically diverse group of fishes in the Lake. We analyzed complete mitochondrial genomes from four species of the endemic Baikal genus Batrachocottus (B. baicalensis, B. multiradiatus, B. talievi, and B. nikolski). Mitogenome sequences are 16,523–16,535 bp in length with a mitogenomic organization and gene arrangement identical to that of typical teleosts. Phylogenetic analysis using the Bayesian method positioned B. baicalensis outside the monophyletic clades of the genus Batrachocottus. Batrachocottus multiradiatus and B. talievi are sister species.
The endemic flock of Baikal sculpins (Cottoidei), according to the most recent calculation, includes 33 species (Sideleva Citation2003). Different species of this unique group are adapted to various depth habitats and diets, subdividing into three ecological groups: benthic, benthopelagic, and pelagic. The genus Batrachocottus (Big-headed sculpins) comprises four species: B. baicalensis (Dybowski, 1874), B. nikolskii (L. S. Berg, 1900), B. multiradiatus (L. S. Berg, 1907) and B. talievi (Sideleva, 1999). Molecular data confirmed the validity of two species within the genus (Kontula et al. Citation2003): B. nikolskii and B. multiradiatus. Here, we analyze for the first time molecular phylogeny involving the B. baicalensis and B. talievi. Among four Batrachocottus species only B. baicalensis, which mainly inhabited coastal shallow terrace zone, can be clearly defined morphologically. Three other species, which occur mostly in the deep-water zone, are often difficultly to identify because they form a «chain of forms» owing to the overlapped variation of morphometric characters (Bogdanov Citation2017). The questions about species distribution, their number, and population structure are still open. The mitochondrial genome-based phylogenetic analysis would improve our understanding of the evolutionary taxonomy and relationship between Batrachocottus species and other Cottoidei.
Here, we sequenced and analyzed the complete mitochondrial DNA sequences of four species of the genus Batrachocottus. All the samples were collected in Lake Baikal by nets and trawl (53°50′27′′N, 108°34′42′′E for B. baicalensis, 55°06′13′′N, 109°40′14′′E for B. multiradiatus, 54°48′30′′N, 109°28′81′′E for B. nikolskii and 53°41′68′′N, 108°38′41′′E for B. talievi). The total genomic DNA was isolated from fin clips by the conventional phenol-chloroform method (Sambrook et al. Citation1989).
Mitogenomes were generated using traditional Sanger sequencing on an ABI 3500 Genetic Analyzer (Applied Biosystems, USA). Primers were designed according to the conserved regions of the complete mitogenomes of other cottoid species. Sequence reads were assembled by CAP3 software (Huang and Madan Citation1999) and were aligned within the ClustalW program in BioEdit software. The lengths of mitogenomes are 16,523 bp for B. baicalensis with 47.67% GC content, 16,530 for B. talievi with 47.57% GC content, 16,532 for B. multiradiatus with 47.70% GC content, and 16,535 bp for B. nikolskii with 47.58% GC content. Mitochondrial genomes contain 2 ribosomal RNA genes, 22 transfer RNA genes, 13 protein-coding genes, and a control region with the gene order and codon usage identical to that of typical teleosts. The part of mitochondrial DNA of B. multiradiatus had a single nucleotide deletion in ND6, resulting in a frameshift. These can be mitochondrial heteroplasmy or nuclear insertion of mitochondrial pseudogenes (numts).
Phylogenetic analysis was performed using the sequences of 13 mitochondrial protein-coding genes of Batrachocottus and those of other Cottoidei species. Optimum evolutionary models of nucleotide substitutions for multilocus phylogeny were selected in PartitionFinder2 (Lanfear et al. Citation2017). The phylogenetic tree () was generated with the Bayesian inference method implemented in MrBayes v3.2.7 (Ronquist et al. Citation2012).
Figure 1. Multilocus Bayesian DNA phylogeny of the Cottoidei based on the nucleotide sequences of the 13 mitochondrial protein-coding genes. Posterior probabilities with 200,000 generations were shown next to nodes.
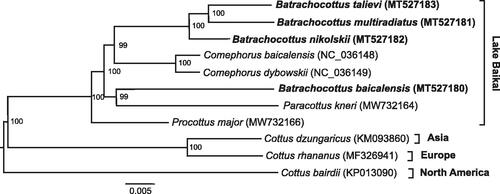
The monophyly of the genus Batrachocottus is not supported by the phylogenetic tree. Only three species, – B. multiradiatus, B. nikolskii, and B. talievi, formed monophyletic clade with bootstrap value = 100%, whereas B. baicalensis is outside the group. The phylogenetic analysis indicated that B. multiradiatus and B. talievi are sister species. This work confirmed the validity of four species, but there was a question about the belonging of the B. baicalensis to the genus Batrachocottus.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank (https://www.ncbi.nlm.nih.gov/) under the accession numbers MT527180 (B. baicalensis), MT527181 (B. multiradiatus), MT527182 (B. nikolskii), and MT527183 (B. talievi).
Specimens were deposited at Limnological Institute of the SB RAS, Irkutsk, Russia (http://lin.irk.ru , contact person: Veronika Teterina, email: [email protected]) under the voucher number Bb20 for B. baicalensis, Bm2_O for B. multiradiatus, Bn316 for B. nikolskii and Bt268 for B. talievi.
Additional information
Funding
References
- Bogdanov BE. 2017. Phenetic relations and problem of species identification of the genus Batrachocottus (Pisces: Cottidae). Hydrob J. 53(1):41–49.
- Huang X, Madan A. 1999. CAP3: a DNA sequence assembly program. Genome Res. 9(9):868–877.
- Kontula T, Kirilchik SV, Vainola R. 2003. Endemic diversification of the monophyletic Cottoid fish species flock in Lake Baikal explored with mtDNA sequencing. Mol Phylogenet Evol. 27(1):143–155.
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34(3):772–773.
- Ronquist F, Teslenko M, van der Mark P, Ayres D, Darling A, HÖhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory.
- Sideleva VG. 2003. The endemic fishes of Lake Baikal. Leiden (The Netherlands): Backhuys; p. 270.
