Abstract
The complete chloroplast and mitochondrial genome sequences of Scopelophila cataractae (Pottiaceae, Bryophyta) are determined. The chloroplast genome is 122,290 bp with 118 genes and the mitochondrial genome is 105,607 bp with 67 genes, both genomes are circular. This study showed the S. cataractae plastome contains the smallest genome size, and a functional trnPGGG gene, relative to other pottiaceous species. Phylogenetic inferences support the sister relationship of S. cataractae to all other pottiaceous accessions.
Pottiaceae is one of the richest families of Bryophyta, comprising more than 1,400 species in approximately 80 genera (Zander Citation1993). Scopelophila cataractae (Mitt.) Broth. is well known as a ‘copper moss’ that can survive in copper-rich environments, such as around temples and shrines with copper roofs, bronze artifacts, abandoned mines, and smelters (Satake et al. Citation1988; Satake Citation2014). The species accumulate copper in the cell wall pectin (Konno et al. Citation2010), although the molecular mechanisms of accumulation have not been revealed. In vascular plants, metal tolerance and homeostasis are maintained by chelating, effluxing, or sequestering molecules (Clemens Citation2001; Hall Citation2002). In addition to the unique ecological features of S. cataractae, molecular phylogenetic studies have revealed that it was included in the basalmost clade within the Pottiaceae (Werner et al. Citation2004; Cox et al. Citation2010; Inoue and Tsubota Citation2016), corresponding to the subfamily Merceyoideae. The genomic resources of organelle have been provided in several genera of the Pottiaceae (Chionoloma: Alonso et al. Citation2016, as Oxystegus; Pseudocrossidium: Cevallos et al. Citation2019, Citation2020; Syntrichia: Oliver et al. Citation2010; Yoon et al. Citation2016; Kim et al. Citation2019). The genomic data are however still limited in the family and lack in the major lineages. Here we present the chloroplast (cp) and mitochondrial (mt) genomes of S. cataractae as the resources for a better understanding of genomic structure and evolutions within Pottiaceae. The phylogenetic relationships within the family were also inferred based on the protein-coding sequences of cp and mt genomes.
Samples of S. cataractae were collected from Tochigi Prefecture, Japan (36°46′16″N 139°42′41″E). A specimen was deposited at the Herbarium of Hiroshima University (HIRO; Director: Tomio Yamaguchi, [email protected]) under the voucher number Y. Inoue 4216. The total DNA was extracted from the axenic in vitro culture of protonemal filaments derived from a single spore (Takio Citation1995, modified), with NucleoSpin Plant II (Macherey–Nagel, Duren) following the manufacturer’s protocols, and sequenced using the Illumina MiSeq platform. A total of approximately 693 K raw reads was analyzed, comprising an average fragment length of 150 bp. Low-quality reads (<Q30), abnormal short reads (<20 bp), and adapter sequences were trimmed using fastp 0.20.0 (Chen et al. Citation2018). After quality control, the GetOrganelle 1.7.1 (Jin et al. Citation2020) was used to assemble the filtered reads with the seed reads comprising published organelle genome sequences of mosses, and the assembled sequences were polished by Pilon 1.23 (Walker et al. Citation2014). The polished sequences were annotated using GeSeq 2.03 (Tillich et al. Citation2017) and manually corrected using the SnapGene 5.2.3 (from GSL Biotech; snapgene.com). Since the chloroplast sequence still contained one gap, the specific primers were designed to bridge the gap with PCR amplification and conventional Sanger sequencing by ABI 3730xl. The final annotated cp and mt sequences were submitted to the DNA Data Bank of Japan (DDBJ) and assigned accession numbers LC634773 for cp and LC634774 for mt.
Phylogenetic analyses were conducted with protein-coding sequences of chloroplast (79 genes) and mitochondrial (38 genes) genomes, respectively. Each data matrix consists of representative species selected from major lineages of mosses based on Liu et al. (Citation2019), including all pottiaceous accessions. Sequences were aligned using MAFFT 7.475 (Katoh and Standley Citation2013), with some manual adjustments by the sequence editor of MEGA 7.0.26 (Kumar et al. Citation2016). Start and stop codons were removed, and gaps were treated as missing data. Kakusan4 (4.0.2016.11.07; Tanabe Citation2011) was used to determine the appropriate substitution model and partitioning scheme for our data based on the corrected Akaike information criterion (AICc: Sugiura Citation1978). RAxML 8.2.9 (Stamatakis Citation2014) was used for maximum likelihood inference using GTR + Γ model, with a rapid bootstrap analysis of 1,000 replicates.
Scopelophila cataractae had a 122,290 bp circular chloroplast genome, which is the smallest among the published cp genomes of the Pottiaceae. The cp genome had a GC content of 28.05% and a typical quadripartite structure, consisting of a large single-copy (LSC) region of 83,728 bp, a small single-copy (SSC) region of 18,620 bp, and a pair of inverted repeats (IRs) of 9,971 bp. It contained 118 genes, including 82 protein-coding genes, 32 tRNA genes, and four rRNA genes. Among the published cp genomes of the Pottiaceae, trnPGGG gene is absent or pseudogenized, while in the S. cataractae, this gene appears to be functional according to the tRNA structure prediction by tRNAscan-SE 2.0 (Lowe and Chan Citation2016). The mitochondrial genome was circular, with 105,607 bp, and a GC content of 39.11%. It contained 67 genes, including 40 protein-coding genes, 24 tRNAs, and three rRNAs.
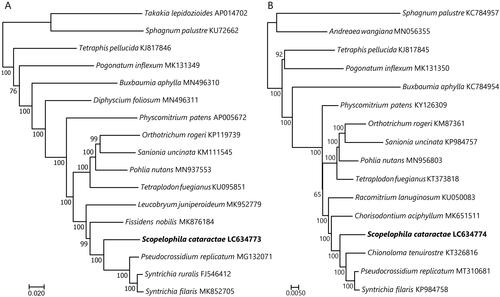
Both cp and mt trees strongly supported the sister relationship of S. cataractae to all other pottiaceous accessions (). This result is consistent with previous phylogenies based on the selected cp genes (Werner et al. Citation2004; Cox et al. Citation2010; Inoue and Tsubota Citation2016).
Figure 1. Maximum-likelihood trees of mosses inferred from the 79 chloroplast (A) and 38 mitochondrial (B) protein-coding sequences. Bootstrap values of 1,000 replicates by RAxML are shown on the branches. The root is arbitrarily placed on the branch leading to the clades that include members of the genera Sphagnum, and Takakia or Andreaea.
Acknowledgments
We express our sincere thanks to Mr. Takayuki Kamiyama for supporting our field research; Ms. Phan Quynh Chi for supporting English translation; and Dr. Misao Itouga and Dr. Toshihisa Nomura for providing valuable information on metal tolerance in plants. MiSeq sequencing and part of data analyses were carried out at the Oral Microbiome Center (Kagawa, Japan), and Sanger sequencing was carried out at Macrogen Japan (Tokyo, Japan). We would like to thank Editage (www.editage.com) for English language editing.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in DDBJ (https://www.ddbj.nig.ac.jp/) and NCBI (https://www.ncbi.nlm.nih.gov/), under the accession numbers LC634773 and LC634774. The associated BioProject, BioSample, and SRA numbers are PRJDB11883, SAMD00334531, and DRR303909 respectively.
Additional information
Funding
References
- Alonso M, Medina R, Cano MJ, Jiménez JA, Goffinet B. 2016. The complete mitochondrial genome of the moss Oxystegus tenuirostris (Hook. & Taylor) A.J.E. Sm. (Pottiaceae, Bryophyta). Mitochondrial DNA A DNA Mapp Seq Anal. 27(5):3808–3809.
- Cevallos MA, Guerrero G, Ríos S, Arroyo A, Villalobos MA, Porta H. 2019. The chloroplast genome of the desiccation-tolerant moss Pseudocrossidium replicatum (Taylor) R.H. Zander. Genet Mol Biol. 42(2):488–493.
- Cevallos MA, Guerrero G, Ríos S, Arroyo A, Villalobos MA, Porta H. 2020. The mitogenome of Pseudocrossidium replicatum, a desiccation-tolerant moss. Mitochondrial DNA B Resour. 5(3):2339–2341.
- Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34(17):i884–i890.
- Clemens S. 2001. Molecular mechanisms of plant metal tolerance and homeostasis. Planta. 212(4):475–486.
- Cox CJ, Goffinet B, Wickett NJ, Boles SB, Shaw AJ. 2010. Moss diversity: a molecular phylogenetic analysis of genera. Phytotaxa. 9(1):175–195.
- Hall JL. 2002. Cellular mechanisms for heavy metal detoxification and tolerance. J Exp Bot. 53(366):1–11.
- Inoue Y, Tsubota H. 2016. Systematics of the family Pottiaceae (Bryophyta) with special reference to the familial and subfamilial circumscriptions. Hikobia. 17:117–129.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kim SC, Byun MY, Kim JH, Lee H. 2019. The complete chloroplast genome of an Antarctic moss Syntrichia filaris (Müll.Hal.) R.H. Zander. Mitochondrial DNA B Resour. 4(2):2303–2304.
- Konno H, Nakashima S, Katoh K. 2010. Metal-tolerant moss Scopelophila cataractae accumulates copper in the cell wall pectin of the protonema. J Plant Physiol. 167(5):358–364.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Liu Y, Johnson MG, Cox CJ, Medina R, Devos N, Vanderpoorten A, Hedenäs L, Bell NE, Shevock JR, Aguero B, et al. 2019. Resolution of the ordinal phylogeny of mosses using targeted exons from organellar and nuclear genomes. Nat Commun. 10(1):1485.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
- Oliver MJ, Murdock AG, Mishler BD, Kuehl JV, Boore JL, Mandoli DF, Everett KD, Wolf PG, Duffy AM, Karol KG. 2010. Chloroplast genome sequence of the moss Tortula ruralis: gene content, polymorphism, and structural arrangement relative to other green plant chloroplast genomes. BMC Genomics. 11:143.
- Satake K. 2014. Do-goke no Fushigi [The mystery of copper bryophytes]. 222 pp. Isebu, Tsukuba. (in Japanese)
- Satake K, Shibata K, Nishikawa M, Fuwa K. 1988. Copper accumulation and location in the moss Scopelophila cataractae. J Bryol. 15(2):353–376.
- Stamatakis A. 2014. RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Sugiura N. 1978. Further analysis of the data by Akaike’s information criterion and the finite corrections. Commun Stat Theory Methods. 7(1):13–26.
- Takio S. 1995. A protocol for isolation and culture of callus in bryophytes. Heredity. 49:44–47. (in Japanese)
- Tanabe AS. 2011. Kakusan4 and Aminosan: two programs for comparing nonpartitioned, proportional and separate models for combined molecular phylogenetic analyses of multilocus sequence data. Mol Ecol Resour. 11(5):914–921.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 9(11):e112963.
- Werner O, Ros RM, Cano MJ, Guerra J. 2004. Molecular phylogeny of Pottiaceae (Musci) based on chloroplast rps4 sequence data. Pl Syst Evol. 243(3–4):147–164.
- Yoon Y-J, Kang Y, Kim M-K, Lee J, Park H, Kim JH, Lee H. 2016. The complete mitochondrial genome of an Antarctic moss Syntrichia filaris (Müll.Hal.) R.H. Zander. Mitochondrial DNA A DNA Mapp Seq Anal. 27(4):2779–2780.
- Zander RH. 1993. Genera of the Pottiaceae: mosses of harsh environments. Bull Buffalo Soc Nat Sci. 32:1–378.
