Abstract
In the present study, the complete mitogenome of Cephaloscyllium fasciatum has been sequenced and assembled. The complete mitochondrial genome is 16,509 bp in length and contains 37 mitochondrial genes and a control region as other fishes. The nucleotide composition of C. fasciatum mitogenome showed an anti-G bias and an excess of AT content. Furthermore, maximum-likelihood phylogenetic tree was constructed based on 13 protein-coding genes of C. fasciatum and other 15 sharks. This work will provide molecular data for studies on phylogeny and evolution in the family Scyliorhinidae.
Introduction
Species of the genus Cephaloscyllium Gill 1862, also known as swellsharks. These species can be distinguished from other sharks in the family Scyliorhinidae due to the lack of labial furrows. Furthermore, the Cephaloscyllium has a unique ability that they could swallow either seawater or air, to inflate their stomachs to deter predation (Schaaf-Dasilva and Ebert Citation2008). Among these species, reticulated swellshark (Cephaloscyllium fasciatum Chan, 1966) is one of the two species distributed in the South China Sea (Nakaya et al. Citation2013). Although the C. fasciatum is a common species in the catch of trawl fishery, studies on this species are insufficient. Therefore, we sequenced the complete mitogenome sequence and performed the phylogenetic analysis of C. fasciatum, the results of the present study would provide basic information for further studies about the species.
Materials and methods
The C. fasciatum specimen was collected from the Beibu Gulf of the South China Sea (N 107.8150°, E 19.2510°) on 9th September 2019. The specimen was identified based on morphological characters followed the identification keys of Nakaya et al. (Citation2013). Then, muscle from the C. fasciatum specimen was collected and frozen in liquid nitrogen for DNA extraction. The sample was stored in the Key Laboratory of South China Sea Fishery Resources Exploitation & Utilization, Ministry of Agriculture Rural Affairs (Binbin Shan, [email protected]) under the voucher number CF_Beibu01. Total genomic DNA was extracted using the Aidlab Genomic DNA Extraction Kit. The genome of C. fasciatum was sequenced by next-generation sequencing (Illumina HisSeq 4000). After trimming, 30.82 million clean reads were obtained. The clean reads were preliminarily assembled using de novo assembly by using SPAdes version 3.10.1 with k-mer size 127 (Bankevich et al. Citation2012). Then, we compared the assembled mitogenome with sequences of C. umbratile (NCBI accession no. NC_029399.1) to check the accuracy of the assembled mitochondrial genome. Furthermore, the mitogenome was annotated by MITOS2 (Bernt et al. Citation2013). Phylogenetic analysis was performed based on the protein-coding genes nucleotide sequences using the iq-tree.
Results
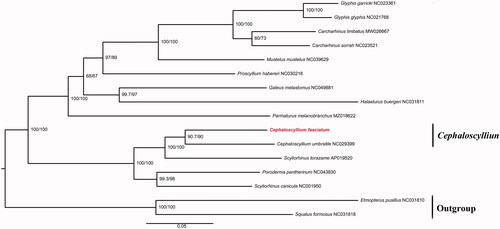
The mitogenome of C. fasciatum is 16,703 bp in length, including a non-coding AT-rich region (D-loop region), 13 protein-coding genes, 2 ribosomal RNA (rRNA) genes, and 22 transfer RNA (tRNA) genes. The 13 protein-coding genes are 11,432 bp, accounting for 68.44% of the whole mitogenome. The nucleotide composition of C. fasciatum mitogenome is estimated to be 31.34% T, 23.58% C, 30.60% A, and 14.48% G, with anti-G bias and an excess of AT as the observation in most fishes (Miya et al. Citation2001). The maximum-likelihood phylogenetic tree was constructed based on 13 protein-coding genes of C. fasciatum and the other 15 sharks using the software iq-tree, Squalus formosus, and Etmopterus pusillus were selected as outgroups (). The most suitable nucleotide sequence model mtART + F + I + G4 was selected through Modeltest based on the Akaike Information Criteria (AIC) (Posada and Crandall Citation1998). As shown in , the evolutionary relationship of C. fasciatum was depicted in the phylogenetic tree which was composed of two main branches. The below branch encompassed C. fasciatum, C. umbratile, and other species in Scyliorhinidae, indicating a closer relationship. Furthermore, the phylogenetic reconstruction supported the sister taxon of C. fasciatum and C. umbratile which conformed to traditional taxonomic estimates (Nakaya et al. Citation2013).
Figure 1. Phylogenetic tree reconstructed by maximum-likelihood (ML) analysis based on 13 protein-coding genes (PCGs) of 16 sharks, including C. fasciatum sequenced (red font) in this study. Numbers below or above branches are assessed by ML bootstrap. Numbers following scientific names are GenBank accessions.
Discussion and conclusion
In the present study, we sequenced and assembled the mitochondrial genome of C. fasciatum. We detected the genes and base compositions of the mitochondrial genome. In addition, to determine the taxonomic status of C. fasciatum, we constructed the phylogenetic tree using the maximum likelihood method on the basis of the 13 protein-coding genes of C. fasciatum and the other 15 species. The phylogenetic tree showed that the C. fasciatum had a closer relationship with species in Scyliorhinidae (). We expect that the results of the present study will facilitate further investigations on the molecular evolution and conservation biology of C. fasciatum.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/nuccore/MZ424309 under accession no. MZ424309.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Miya M, Kawaguchi A, Nishida M. 2001. Mitogenomic exploration of higher teleostean phylogenies: a case study for moderate-scale evolutionary genomics with 38 newly determined complete mitochondrial DNA sequences. Mol Biol Evol. 18(11):1993–2009.
- Nakaya K, Inoue S, Ho H-C. 2013. A review of the genus Cephaloscyllium (Chondrichthyes: Carcharhiniformes: Scyliorhinidae) from Taiwanese waters. Zootaxa. 3752(1):101–129.
- Posada D, Crandall KA. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics. 14(9):817–818.
- Schaaf-Dasilva JA, Ebert DA. 2008. A re-description of the eastern Pacific swellshark, Cephaloscyllium ventriosum (Garman 1880) (Chondrichthyes: Carcharhiniformes: Scyliorhinidae), with comments on the status of C. uter (Jordan & Gilbert 1896). Zootaxa. 1872(1872):59–68.
