Abstract
We sequenced and annotated the complete mitochondrial genome of Ctenochaetus tominiensis (Randall Citation1955) from Indonesia. The genome was assembled into a circular molecule of 16,429 bp with 44.45% GC content. This genome consisted of 13 protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes, and 1D-loop. Phylogenetic analysis based on 13 PCGs showed that Ctenochaetus and Acanthurus were recovered in a single clade. The mitochondrial genome of C. tominiensis is helpful for species identification and phylogenetic position of fish.
Ctenochaetus tominiensis (Randall Citation1955) is a widely distributed fish species in the Western Pacific, Indonesia, the Philippines, New Zealand, and is mostly found around coral reef. This fish species is attributed to Acanthuridae, Acanthurinae, Ctenochaetus. Ctenochaetus tominiensis has a thick-walled stomach similar to Acanthurus (Clements et al. Citation2003). Analysis on the complete mitochondrial genome of fish will provide the basis for species identification and biological diversity (Sorenson et al. Citation2013).
Ctenochaetus tominiensis was collected from Bali, Indonesia (115°2′E, 8°7′S). A specimen was deposited at Guangdong Ocean University (Qingzhu Chu, [email protected]) under the voucher number hdfh00685. Genomic DNA was extracted with DNAzol™ Reagent from muscle tissue of three C. tominiensis. DNA was used to construct the library for sequencing. The mitochondrial genome was sequenced using Illumina HISeq PE150 platform (Sangon Biotech, Shanghai, China). BBMap software (https://escholarship.org/uc/item/1h3515gn) was used to process raw data. The mitochondrial genome was assembled and annotated using NOVOPlasty (Dierckxsens et al. Citation2017) and MITOS2 software (Gomes de Sá et al. Citation2017) with A. leucosternon EU136032 as reference. The complete mitochondrial genome of C. tominiensis is circular and 16,429 bp long. The base composition of this mitochondrial genome is 29.22%A, 26.33%T, 28.60%C, and 15.85%G. The mitochondrial genome contains 13 protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes, and one non-coding region (D-loop).
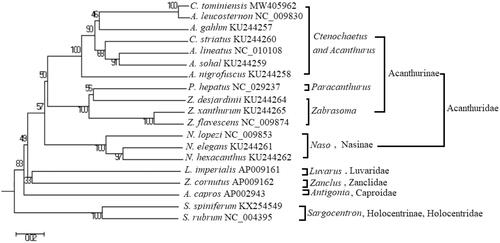
The phylogenetic position of C. tominiensis was analyzed by phylogenetic trees based on 13 PCGs of 19 fishes, with Sargocentron as the outgroup. All sequences were aligned using DNAstar software, and evolutionary models of 13 PCGs were simulated using ModelTest3.8 software. Neighbor-joining, maximum parsimony (MP), minimum evolution, and UPGMA trees were constructed using MEGA6.0 software. MrBayes tree was constructed using MrBayes3.12 software with the general time-reversible + gamma + invariants model. The MP trees were also built using PAUP4.10 by running the heuristic search. The topological structures of all phylogenetic trees were basically similar (). Ctenochaetus tominiensis with other species of Ctenochaetus and Acanthurus were recovered in a single clade, which is consistent with other studies (Sorenson et al. Citation2013; Grulois et al. Citation2020). The phylogenetic position of other basically coincided with traditional taxonomy. Ctenochaetus and Acanthurus should be synonymized when evolution studied.
Disclosure statement
No potential conflict of interest was reported by authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/nuccore/MW405962 under the accession no. MW405962. The associated BioProject, Bio-Sample, and SRA numbers are PRJNA718725, SAMN18559513, and SRR14163267, respectively. Mitochondrial sequences of Acanthurus leucosternon (NC_009830), Acanthurus lineatus (NC_010108), Acanthurus shoal (KU244259), Acanthurus nigrofuscus (KU244258), Acanthurus gahhm (KU244257), Ctenochaetus striatus (KU244260), Paracanthurus hepatus (NC_029237), Zebrasoma flavescens (NC_009874), Zebrasoma xanthurum (KU244265), Zebrasoma desjardinii (KU244264), Naso lopezi (NC_009853), Naso hexacanthus (KU244262), Naso elegans (KU244261), Luvarus imperialis (AP009161), Zanclus cornutus (AP009162), Antigonia capros (AP002943), Sargocentron rubrum (NC_004395), and Sargocentron spiniferum (KX254549) are open at https://www.ncbi.nlm.nih.gov.
Additional information
Funding
References
- Clements KD, Gray RD, Howard Choat J. 2003. Rapid evolutionary divergences in reef fishes of the family Acanthuridae (Perciformes: Teleostei). Mol Phylogenet Evol. 26(2):190–201.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Grulois D, Hogan RI, Paygambar S, Planes S, Fauvelot C. 2020. New microsatellite DNA markers to resolve population structure of the convict surgeonfish, Acanthurus triostegus, and cross-species amplifications on thirteen other Acanthuridae. Mol Biol Rep. 47(10):8243–8250.
- Gomes de Sá P, Veras A, Fontana CS, Aleixo A, Burlamaqui T, Mello CV, de Vasconcelos ATR, Prosdocimi F, Ramos R, Schneider M, et al. 2017. The assembly and annotation of the complete Rufous-bellied thrush mitochondrial genome. Mitochondrial DNA Part A. 28(2):231–232.
- Randall JE. 1955. A revision of the surgeon fish genus Ctenochaetus, family Acanthuridae, with descriptions of five new species. Zoologica. 40(15):149–166.
- Sorenson L, Santini F, Carnevale G, Alfaro ME. 2013. A multi-locus timetree of surgeonfishes (Acanthuridae, Percomorpha), with revised family taxonomy. Mol Phylogenet Evol. 68(1):150–160.