Abstract
Prunus clarofolia (Schneid.) Yu et Li is a very attractive wild flowering cherry endemic to China. In this study, the complete chloroplast genome of P. clarofolia was assembled. The total length of the chloroplast genome was 157,899 bp, containing a pair of inverted repeat regions of 26,393 bp each, separated by a small single-copy region of 19,142 bp, and a large single-copy region of 85,971 bp. The overall GC content of the chloroplast genome was 36.71%. The genome contained 131 genes, including 85 protein-coding genes, 37 tRNA genes, eight rRNA genes, and one pseudogene. Phylogenetic analysis revealed that P. clarofolia and P. pseudocerasus showed the closest relationship.
Prunus clarofolia (Schneid.) Yu et Li (Rosaceae), also named Cerasus clarofolia, is a deciduous shrubs or small trees native to China. It is mainly distributed in Anhui, Gansu, Guizhou, Hebei, Henan, Hubei, Hunan, Yunnan, and Zhejiang province, and usually grows in forests or thickets on mountain slopes at an altitude of 800–3600 m (Li and Bartholomew Citation2003). A good knowledge of genome information and the phylogenetic position of P. clarofolia would contribute to the formulation of efficient strategies for its conservation, management, and utilization. Here, we reported the complete chloroplast genome of P. clarofolia and ascertained its phylogenetic position.
The fresh leaves of P. clarofolia were collected from Tianmen Mountain in Zhangjiajie city, China (29°3′N, 110°28′E). A specimen was deposited at Herbarium (PE), Institute of Botany, CAS Flora of China (https://www.cvh.ac.cn/spms/detail.php?id=e6f99a52, Caifei Zhang, [email protected]) under the voucher number 02247373, and sequenced DNA can be found in the laboratory of Hunan Botanical Garden (voucher specimen: PCHN2020-5, HBG, 113°2′18.44″,28°6′34.54″). Total genomic DNA was extracted by modified CTAB protocol (Doyle and Doyle Citation1987). The whole genome was sequenced with the Illumina NovaSeq 6000 Sequencing System following the manufacturer’s protocol (Illumina, San Diego, CA). A total of 5.4 Gb clean data were obtained and then assembled using SPAdes version V 3.10.1 (Bankevich et al. Citation2012). Finally, the genome assemblies were annotated by using CPGAVAS2 (Shi et al. Citation2019), followed by manual correction.
The chloroplast genome of P. clarofolia (accession number: MT747185) was 157,899 bp in length and contained a pair of inverted repeat regions (IRs) of 26,393 bp each, a small single-copy (SSC) region of 19,142 bp, and a large single-copy (LSC) region of 85,971 bp. The overall GC content of the chloroplast genome was 36.71%, and the values of the LSC, SSC, and IR regions were 34.59%, 30.16%, and 42.53%, respectively. The chloroplast genome contained 131 genes, including 85 protein-coding genes, 37 tRNA genes, eight rRNA genes, and one pseudogene.
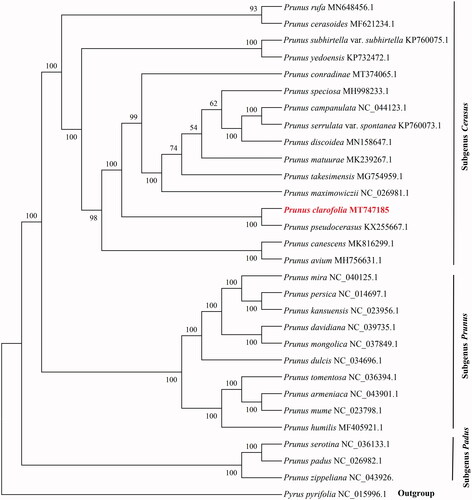
We used the whole chloroplast genome sequences of 28 Prunus species to ascertain the phylogenetic position of P. clarofolia. Pyrus pyrifolia (NC_015996) was used as the outgroup. The best-fit model generalized time-reversible (GTR) for DNA sequence evolution was selected using jModelTest v2.1.10 (Darriba et al. Citation2012) based on Akaike’s information criterion. The phylogenetic tree was constructed with 1000 bootstrap replicates under the maximum-likelihood (ML) inference by using RAxML v8.2.10 (Miller et al. Citation2010). The ML tree revealed that all the species of Prunus were clustered into three clades (bootstrap support value 100%), subgenus Cerasus, subgenus Prunus, and subgenus Padus (). This result is consistent with the previous studies (Chin et al. Citation2014; Yan et al. Citation2020). In the tree, P. clarofolia and P. pseudocerasus showed the closest relationship.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession number MT747185. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA739540, SRR14885714, and SAMN19791814, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Chin SW, Shaw J, Haberle R, Wen J, Potter D. 2014. Diversification of almonds, peaches, plums and cherries – molecular systematics and biogeographic history of Prunus (Rosaceae). Mol Phylogenet Evol. 76:34–48.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Li CL, Bartholomew B. 2003. Cerasus. In: Wu ZY, Raven PH, editors. Flora of China. Vol. 9. Beijing/St. Louis: Science Press/Missouri Botanical Garden Press; pp. 404–420.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Gateway Computing Environments Workshop 14; p. 1–8.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyser. Nucleic Acids Res. 47:65–73.
- Yan JW, Li JH, Bai WF, Yu L, Nie DL, Xiang ZH, Wu SZ. 2020. The complete chloroplast genome of Prunus conradinae (Rosaceae), a wild flowering cherry from China. Mitochondrial DNA B Resour. 5(3):2153–2154.