Abstract
Xenopsylla cheopis, also called oriental rat flea, is an ectoparasite as well as disease vector for murine typhus and bubonic plague. In the study, the whole mitochondrial genome of X. cheopis was sequenced and assembled, which is the second report of mitochondrial genome in the family Pulicidae and the sixth mitochondrial genome in the order Siphonaptera (fleas). The mitochondrial genome is 18,902 bp in length, consisting of 40% A, 44% T, 6% G, and 10% C. Phylogenetic analysis of all available mitochondrial genomes from Siphonaptera indicated that X. cheopis clustered with Ctenocephalides felis since both species belonged to the family Pulicidae. The complete mitochondrial genome of X. cheopis could serve as useful genetic data for investigating the genetic relationship of fleas.
Xenopsylla cheopis (Rothschild, 1903) (Siphonaptera: Pulicidae), common name of oriental rat flea, is an ectoparasite as well as disease vector. X. cheopis is typically brown color and grows to be approximately 2.5 mm in length. The morphological features of X. cheopis are different from cat fleas (Ctenocephalides felis) and dog fleas (Ctenocephalides canis) by lacking of a pronotal comb, genal comb or divided mesopleuron (Wells and Elston Citation2020).
The hosts of X. cheopis are mammals, primarily Rattus species, such as Rattus tanezumi and Rattus norvegicus (Xie Citation1981). Human is an incidental host of X. cheopis. If X. cheopis leaves the host, it can survive up to six weeks before migrating to a new host (Slavicek Citation2008). The host switching among sympatric species increases the chance of disease transmission. X. cheopis transmits pathogens including Rickettsia typhi and Yersinia pestis, which can cause murine typhus and bubonic plague, respectively. R. typhi in the feces of fleas might enter human body by aerosol inhalation, and infect mainly endothelial cells thought the body causing interstitial pneumonia, meningoencephalitis, fever, etc. Plague is the most severe disease spread by X. cheopis that can transmit plague pathogen Y. pestis to a new host after feeding on a previous host with bacteremia. After establish infection, Y. pestis survives in the gut of fleas and might return to the bite site as the blood meal regurgitates, which could lead to an alternative mode of transmission (Bacot and Martin Citation1914). The flea-borne transmission of Y. pestis to human can be achieved by fleabite. Besides pathogen transmission, the bite of X. cheopis itself causes pruritic lesions on human skin. As global warming situation continues, increased population of X. cheopis could be an emerging public health issue (Wells and Elston Citation2020).
The understanding of X. cheopis mitochondrial genome (mitogenome) could enrich our knowledge on phylogenetic relation of species from the order Siphonaptera (fleas) that comprises 246 genera (Lewis Citation1998). However, there are only six mitogenomes from Siphonaptera in GenBank. In the study, the whole mitogenome of X. cheopis was sequenced, assembled and annotated, which was the second mitogenome reported in the family Pulicidae.
The X. cheopis specimens used in this study were isolated from a trapped house rat (R. tanezumi) in Shanghai, China (N31°13′, E121°29′). Collected specimens were identified morphologically and stored in alcohol at 4 °C. A specimen was deposited at National Institute of Parasitic Diseases (en.ipd.org.cn, Contact: Chunhua Gao, email: [email protected]) under the voucher number IPD2020XC01. The total DNA of each rat flea was extracted using QIANamp Micro DNA Kit (Qiangen, Germantown, MD, USA). The sequencing of X. cheopis mitogenome was performed on an Illumina HiSeq 2500 system with a paired-end 150 bp sequencing strategy. The mitogenome was assembled using MitoZ v2.3 (Meng et al. Citation2019). Gene boundaries were identified by MITOS2 Web Server (Bernt et al. Citation2013). The protein-coding genes (PCGs) and rRNA genes were verified through homology comparison with those of C. felis. The tRNA genes were verified using tRNAscan-SE version 2.0 (Chan and Lowe Citation2019).
The mitogenome of X. cheopis is 18,902 bp in length (GenBank accession No. MW310242), consisting of 40% A, 44% T, 6% G and 10% C, which contains 13 PCGs, 22 tRNA genes, 2 rRNA genes, and an AT-rich control region. The gene arrangement follows the typical order of metazoan gene set (Cameron Citation2014). Most of the PCGs utilize ATA as their start codon except cox3, nad1, nad4, and nad5 which use ATG. All PCGs ended with a TAA stop codon. For cox1, cox2, nad3, nad4, nad4l, and nad5, TAA stop codon is completed by the addition of 3′ A to the mRNA.
The mitogenome of X. cheopis is most closed to that of C. felis by comparison with available mitogenomes (Driscoll et al. Citation2020). Both C. felis and X. cheopis are under the family Pulicidae. Based on the sequences of 13 PCGs in mitogenome, a phylogenetic tree was constructed using maximum likelihood (ML) method through MEGA X using GTR G + I substitution model (Kumar et al. Citation2018). Siphonaptera was genetically closed to Mecoptera (scorpionflies), both of which belong to Superorder Endopterygota (Holometabola) (Whiting Citation2002). Therefore, in the phylogenetic analysis of Siphonaptera, the species from Mecoptera were set as outgroups ().
Figure 1. The maximum likelihood tree of six species from Siphonaptera is based on 13 protein-coding genes in mitochondrial genomes. Bootstrap values in percentage are shown at nodes. Four species from Mecoptera were set as outgroups. GenBank accession numbers are listed following species name.
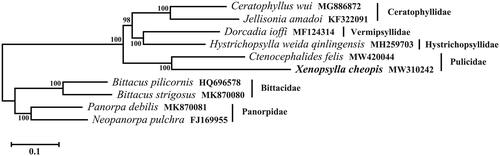
In the phylogenetic tree, X. cheopis and C. felis clustered together, forming the branch of family Pulicidae. Ceratophyllus wui and Jellisonia amadoi belonging to the family Ceratophyllidae formed one cluster. Bittacus strigosus, Bittacus pilicornis, Panorpa debilis, and Neopanorpa pulchra which belong to the order Mecoptera separated clearly from the species of Siphonaptera. The branches were well supported. In conclusion, the complete mitochondrial genome of X. cheopis provided useful genetic data for investigating the genetic relationship of fleas.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov (GenBank: MW310242 BioProject: PRJNA754093, BioSample: SAMN20741547, SRA: SRR15425125).
Additional information
Funding
References
- Bacot AW, Martin CJ. 1914. LXVII. Observations on the mechanism of the transmission of plague by fleas. J Hyg. 13(Suppl):423–439.
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Cameron SL. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59:95–117.
- Chan PP, Lowe TM. 2019. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol Biol. 1962:1–14.
- Driscoll TP, Verhoeve VI, Gillespie JJ, Johnston JS, Guillotte ML, Rennoll-Bankert KE, Rahman MS, Hagen D, Elsik CG, Macaluso KR, Azad AF. 2020. A chromosome-level assembly of the cat flea genome uncovers rampant gene duplication and genome size plasticity. BMC Biol. 18(1):70.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Lewis RE. 1998. Résumé of the Siphonaptera (Insecta) of the world. J Med Entomol. 35(4):377–389.
- Meng GL, Li YY, Yang CT, Liu SL. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Slavicek LC. 2008. The Black Death. New York: Chelsea House Publications.
- Wells LE, Elston DM. 2020. What's eating you? Oriental rat flea (Xenopsylla cheopis). Cutis. 106(3):124–126.
- Whiting MF. 2002. Mecoptera is paraphyletic: multiple genes and phylogeny of Mecoptera and Siphonaptera. Zool Scripta. 31(1):93–104.
- Xie BQ. 1981. Ecological habits of Xenopsylla cheopis. Chin J Appl Entomol. 5:47–48.
