Abstract
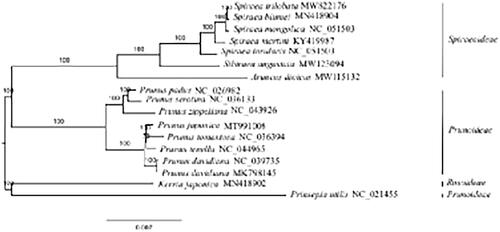
The complete plastome of Spiraea trilobata, a shrub, is determined. The plastome is 155,981 bp in length and comprises a large single-copy region (84,417 bp), a small single-copy region (18,878 bp), and a pair of inverted repeats regions (26,343 bp). A total of 113 unique genes are annotated for the plastome of S. trilobata, containing 79 protein coding genes (PCGs), 30 tRNAs, and four rRNAs. The GC content of this plastome is 36.8%. Phylogenomic analysis based on 17 plastomes reveals that S. trilobata is sister to Spiraea blumei. Compared with S. blumei, S. trilobata lacks both trnM-CAU and ycf1. In addition, the GC content of both species is the same.
S. trilobata belongs to Spiraea in Spiraeoideae of Rosaceae. It is mainly distributed in China, Russia and Turkey (Yu and Kuan Citation1963) and often grows in the shrubs on rocky slopes, forest margins, roadsides and ditches with an altitude of 450–2400 m (Huo et al. Citation2019). It has strong adaptability and is the dominant species in poor soil (Liu et al. Citation2017). It has beautiful flowers and delicate leaves, so it is a common ornamental shrub in garden and has important horticultural and economic value (Yu et al. Citation2018). The leaves and fruits of S. trilobata have the functions of promoting blood circulation, removing blood stasis, detumescence and relieving pain (Olennikov and Chirikova Citation2018). However, the complete plastome of S. trilobata has not been reported. In this study, we showed that the plastome of S. trilobata, which would be helpful for species identification and the phylogenetic analysis of the Rosaceae.
Silica-dried leaves of S. trilobata were collected from Lushan Forest Park (Shandong, China; 23°32′ N, 118°6′ E). The voucher specimen (XLC47) was deposited at College of Life Sciences, Shandong Normal University. Modified CTAB method was used for plant total DNA extraction (Doyle and Doyle Citation1987; Guo et al. Citation2020). The total genomic DNA was used for library preparation and paired-end (PE) sequencing by the Illumina MiSeq instrument at Novogene (Beijing, China). The plastome was assembled using getorganelle (Jin et al. Citation2020). Annotation was performed with PGA-Plastid Genome Annotator (Qu et al. Citation2019), coupled with manual correction using Geneious v8.0.2 (https://www.geneious.com). To determine the phylogenetic placement of S. trilobata, a maximum likelihood (ML) tree was reconstructed using RAxML v8.2.10 (Stamatakis Citation2014), including tree robustness assessment using 1000 rapid bootstrap replicates with the GTRGAMMA substitution model, based on the alignment of 79 shared PCGs using MAFFT v7.313 (Katoh and Standley Citation2013).
The complete plastome of S. trilobata (GenBank accession number: MW822176) is a circular molecular of 155,981 bp in length, consisting of a large single-copy region (84,417 bp), a small single-copy region (18,878 bp), and a pair of inverted repeats regions (26,343 bp). It encodes 113 unique genes, including 79 PCGs, 30 tRNAs, and four rRNAs. The GC content of this plastome is 36.8%. Phylogenomic analysis based on 17 plastomes reveals that S. trilobata is sister to Spiraea blumei (). By comparing the plastomes of S. trilobata and S. blumei, we find that S. trilobata lacks two genes trnM-CAU and ycf1. The GC content of both species is the same.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MW822176. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA720486, SRR14180363, and SAMN18651951, respectively.
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Guo XX, Dai C, Wang R, Q XJ, Z XJ. 2020. Characterization and phylogenetic analysis of the complete plastome of Alopecurus japonicus (Gramineae), an annual weed. Mitochondrial DNA Part B. 5(1):396–397.
- Huo Y, Yan M, Zhao X, Zhu Z, Yuan Z. 2019. The complete chloroplast genome sequence of Spiraea blumei G. Don (Rosaceae). Mitochondrial DNA Part B. 4(2):3671–3672.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. Getorganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol.21(1):241.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Liu HM, Zhang J, Liu JX, Qing S, Zhang Q, Gao Y, Yang CP. 2017. Study of the physiological mechanisms of two species of Spiraea during adaptation to drought treatment. Acta Physiol Plant. 39(8):186.
- Olennikov DN, Chirikova NK. 2018. Rhamnetin glycosides from the genus Spiraea. Chem Nat Compd. 54(1):41–45.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):1–12.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30 (9):1312–1313.
- Yu SX, Gadagkar SR, Potter D, Xu DX, Zhang M, Li ZY. 2018. Phylogeny of Spiraea (Rosaceae) based on plastid and nuclear molecular data: implications for morphological character evolution and systematics. Pers Plant Ecol Evol Syst. 34:109–119.
- Yu T, Kuan K. 1963. Taxa nova Rosacearum Sinicarum (1) (in Chinese). Acta Phytotaxonomica Sinica. 8:206–236.