Abstract
Manglietia ventii is a highly ornamental timber tree species that is listed as endangered on the IUCN Red List. This study was conducted on the complete chloroplast genome of M. ventii. The length of the chloroplast genome is 159,950 bp with GC content as 39.3%. One hundred and thirty-one functional genes were identified in the genome, which included 86 protein-coding genes (PCGs), 37 tRNA genes, and eight rRNA genes. The phylogenetic analysis indicated that M. ventii is most closely related to M. megaphylla and M. aromatica, and the study provides new insights into the evolution of the Magnoliaceae.
Manglietia ventii is a species of timber with high ornamental value, which belongs to Magnoliaceae. The species is endemic to China and occurs in a small area in southeast Yunnan. M. ventii generally grows in valley woods at 800–1200 m (Flora of China Editorial Committee Citation1996). Due to the over-utilization of resources, the increasing destruction of primary forests, and the decline of the natural reproductive capacity, some Magnoliaceae are on the verge of extinction or even extinct (Cicuzza et al. Citation2007; Sima and Lu Citation2009). M. ventii is listed as an endangered species in the Red List, with no more than 100 adult individuals presumably in the wild (IUCN Citation2014). Here, its complete chloroplast genome was sequenced, assembled, and analyzed, which will provide favorable tools for researching the phylogenetic and origination of different Magnoliaceae.
Mature, healthy, and fresh leaves from M. ventii were collected from Kunming (102°45′E, 25°3′N, 1900 m), Yunnan province of China. The voucher specimen was deposited in the herbarium of Southwest Forestry University (http://www.swfu.edu.cn/, contact person: Jingyu Peng, [email protected], accession number: SWFU-MAG-MMV-7739). (The experimental samples for this study were cultivated materials, collected from the campus of Southwest Forestry University and brought back to the studio for experimental studies, and the experimental materials were collected in accordance with relevant institutional, national and international guidelines and regulations. The samples were collected without causing harm to the plant habitat or the trees themselves). Extraction of total genomic DNA using a modified CTAB method and sequencing by Annoroad Gene Technology (Beijing, China) on Illumina Novaseq 6000 platform (Yang et al. Citation2014). The clean data were annotated in Geneious v8.1.3 software (Geneious, Auckland, New Zealand) using the Manglietia longirostrata (GenBank accession number MT584886) complete chloroplast genome as a reference. GetOrganelle v1.6.0 software (Jin et al. Citation2020) was used to assemble the complete chloroplast genome. The successfully annotated genome was submitted to GenBank (accession number: MW415421).
The complete chloroplast genome is a typical loop-like molecule of a four-part structure of 159,950 bp in length, which contains a small single-copy (SSC) replication region of 18,800 bp and a large single-copy (LSC) replication region of 88,008 bp linked by two inverted repeats regions (IRa and IRb) of 26,571 bp. The overall GC content is 39.3%, while the GC content of SSC, LSC, and IR regions is 34.0%, 38.0%, and 43.0%, respectively. There are 86 protein-coding genes (PCGs), eight rRNA, and 37 tRNA. Among these genes, nine PCGs (petB, atpF, ndhA, two ndhB, rpoC1, two rpl2, and rps16) and all tRNAs contain one intron, whereas four (two rps12, clpP, and ycf3) have two introns.
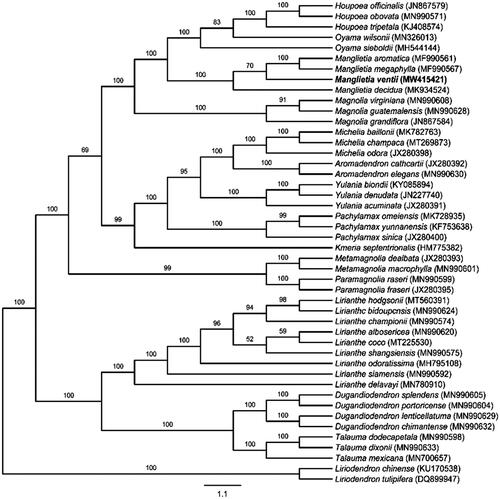
To reveal the evolutionary position of M. ventii among Magnoliaceae, M. ventii and the previously reported 45 cp genomes were aligned to the chloroplast genome matrix using MAFFT v7.487 (Katoh and Standley Citation2013; Chen et al. Citation2020; Sima et al. Citation2021) for ML analysis was based on the TVM + F+R2 model (Kalyaanamoorthy et al. Citation2017), using 1000 bootstrap replicates (Gao et al. Citation2018). Based on the system of Magnoliaceae by Sima and Lu (Citation2012), the phylogenetic analysis indicates that M. ventii is most closely related to M. megaphylla and M. aromatica (). Although M. ventii is separate from M. decidua, they still have a very strong support. Liriodendron chinense and Liriodendron tulipifera of the subfamily Liriodendroideae were served as the outgroup. All genera mentioned for this analysis are monophyletic according to the system of Magnoliaceae by Sima and Lu (Citation2012). These results will provide valuable information for the phylogenetic and evolutionary study of the Magnoliaceae.
Authors contributions
Yuchang Wang: wrote, reviewed, edited the manuscript, and formal analysis. Dawei Wang: funding acquisition, visualization, and project administration. All authors have read and agreed to the published version of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are available in NCBI GenBank (https://www.ncbi.nlm.nih.gov/) under the GenBank accession number MW415421. The associated BioProject, SRA, and BioSample numbers are PRJNA770098, SRR16352012, and SAMN22209219, respectively.
Additional information
Funding
References
- Chen SY, Wu T, Ma HF, Fu YP, Zhu YF, Hao JB, Jiang H, Sima YK. 2020. The complete chloroplast genome sequence of Lirianthe hodgsonii, a tree species of Magnoliaceae as least concern. Mitochondrial DNA B Resour. 5(3):3064–3084.
- Cicuzza D, Newton A, Oldfield S. 2007. The Red List of the Magnoliaceae. Cambridge, UK: Fauna & Flora International; p. 17–42.
- Flora of China Editorial Committee. 1996. Flora of China. Vol. 30. Beijing: Science Press; p. 94.
- Gao F, Shen J, Liao F, Cai W, Lin S, Yang H, Chen S. 2018. The first complete genome sequence of narcissus latent virus from Narcissus. Arch Virol. 163(5):1383–1386.
- IUCN. 2014. Magnolia ventii. The IUCN Red List of Threatened Species. https://doi.org/http://dx.doi.org/10.2305/IUCN.UK.2014-1.RLTS.T191514A1986764.en.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Sima YK, Lu SG. 2009. Magnoliaceae. In: Shui YM, Sima YK, Wen J, Chen WH, editors. Vouchered flora of Southeast Yunnan. Vol. 1. Kunming: Yunnan Science & Technology Press; p. 16–67.
- Sima YK, Lu SG. 2012. A new system for the family Magnoliaceae. In: Xia NH, Zeng QW, Xu FX, Wu QG, editors. Proceedings of the Second International Symposium on the Family Magnoliaceae. Wuhan: Huazhong University Science & Technology Press; p. 55–71.
- Sima YK, Wu T, Chen SY, Ma HF, Hao JB, Fu YP, Zhu YF. 2021. The complete chloroplast genome sequence of Michelia balansae var. balansae (Aug. Candolle) Dandy, a timber and spices species in Magnoliaceae. Mitochondrial DNA B Resour. 6(2):465–467.
- Yang JB, Li DZ, Li HT. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Resour. 14(5):1024–1031.