Abstract
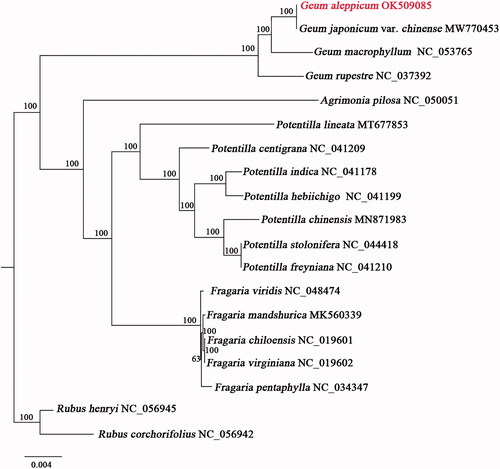
Geum aleppicum is an important landscaping plant with high medicinal value. In this study, the complete chloroplast genome of G. aleppicum was assembled and analyzed. The assembled genome is 156,036 bp in length, which contains a large single-copy (LSC) region of 85,358 bp, a small single-copy (SSC) region of 18,410 bp, and two inverted repeat regions (IRa and IRb) of 26,134 bp. A total of 137 genes were predicted, including 89 protein-coding genes, 40 tRNA genes, and eight rRNA genes. The phylogenetic analysis indicated that G. aleppicum had the closest relationship with G. japonicum var. chinense, and all the species of Geum grouped together with highly bootstrap support.
Geum aleppicum Jacq. is a herbaceous perennial belonged to the family Rosaceae, widely distributed in the temperate, warm temperate and arctic regions of the northern hemisphere (Gajewski Citation1959). It is a traditional Chinese medicinal and edible plant (Li et al. Citation2006). Previous studies had shown that G. aleppicum contains a variety of important active ingredients, such as flavonoids, benzene C, tannins (Yang et al. Citation2013), which have antiviral anti-virus (Xu et al. Citation2000), anti-coagulation (Zeng et al. Citation1998), and anti-oxidation (Choi et al. Citation1997) pharmacological effects. At the same time, as its strong adaptability and easy cultivation, it is often used for rapid greening and landscaping (Song et al. Citation2015). At present, the main research of G. aleppicum was on the chemical composition and efficacy. However, the chloroplast genome information has not been determined. In this study, we reported the complete chloroplast genome of G. aleppicum for the first time using high-throughput sequencing technology, in order to provide a reference for phylogeny and evolution of the genus Geum.
The fresh leaves of G. aleppicum were collected from Nanjing, Jiangsu Province, China (31°54′18″ N, 118°55′5″ E) for DNA extraction. The voucher specimen was deposited at the herbarium of Medicinal Botanical Garden in China Pharmaceutical University (accession number: CPU-LBQ1703, Xu Lu, [email protected]). The whole genomic DNA was extracted using a modified CTAB method (Doyle and Doyle Citation1987) and sequenced on Illumina Hiseq 2500 platform by the Beijing Genomics Institution (BGI). First, raw reads were filtered using Trimmomatic v0.38 (Bolger et al. Citation2014). Next, the filtered clean reads were assembled using NOVOPlasty v2.7.2 (Dierckxsens et al. Citation2016). Using the chloroplast genome of G. rupestre (GenBank accession no. NC_037392) as a reference sequence, the assembled genome of G. aleppicum was annotated by the online tool GeSeq (https://chlorobox.mpimp-golm.mpg.de/geseq.html). Then, the annotated genes were checked and adjusted manually. Finally, we submitted the annotated complete chloroplast genome to NCBI with accession number OK509085.
The complete chloroplast genome of G. aleppicum has a typical tetrad structure with a total length of 156,036 bp. It contains a large single-copy (LSC) region of 85,358 bp, a small single-copy (SSC) region of 18,410 bp, and a pair of inverted repeat regions (IRa and IRb) of 26,134 bp, respectively. There are a total of 137 genes in the chloroplast genome of G. aleppicum, including 89 protein-coding genes, 40 tRNA genes, and eight rRNA genes. Among them, 21 genes contained one intron and two genes (clpP and ycf3) contained two introns. The nucleotide composition is 31.21% A, 18.76% C, 17.99% G, and 32.03% T. The overall GC content is 36.75%, while the GC content of LSC, SSC, and IR regions is 34.45%, 30.74%, and 34.45%, respectively.
We downloaded the sequence data of the chloroplast genomes of 18 closely related species of G. aleppicum from NCBI, and selected the common coding sequences in the chloroplast genomes of these species to construct the phylogenetic tree. Among them, Rubus corchorifolius and Rubus henryi are outgroups. These sequences were aligned using MAFFT v7 (Katoh and Standley Citation2013), and subsequently examined manually by BioEdit v5.0.9 (Hall Citation1999). The maximum-likelihood (ML) phylogenetic tree was constructed using RAxML (Stamatakis Citation2014) with 1000 bootstrap replicates. The result showed that G. aleppicum and G. japonicum var. chinense had the closest relationship, and all Geum species grouped together with highly bootstrap support (). This study will provide a reference for phylogeny and evolution of species of Geum, and be helpful for the further development and utilization of G. aleppicum.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under accession number OK509085. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA770511, SRR16295130, and SAMN22222461, respectively.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Choi YH, Kim MJ, Lee HS, Kwa Hu C, Kwak S. 1997. Antioxidants in leaves of Rosa rugosa. Korean J Pharmacogn. 28(4):179–184.
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):1–9.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Gajewski W. 1959. Evolution in the genus Geum. Evolution. 13(3):378–388.
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment program for Windows 95/98/NT. Nucleic Acids Symp Ser. 41:95–98.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Li g, Yang XH, Wang Y, Mou FH, Dong L, Chen D. 2006. Isolation and structure identification of phenolic compounds from Geum aleppicum Jacq. J Jilin Univ (Med Ed). 32(2):196–198.
- Song JZ, Wang ZX, Zhang YK, Qin JM. 2015. Structure and growth regulation of Geum aleppicum modules of population. Jilin J Tradit Chin Med. 35(12):1252–1254.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Xu HX, Ming DS, Dong H, But PPT. 2000. A new anti-HIV triterpene from Geum japonicum. Chem Pharm Bull. 48(9):1367–1369.
- Yang W, Zheng MS, Lu HZ. 2013. Study the chemical constituents of Geum aleppicum Jacq. J Med Sci Yanbian Univ. 36(1):32–34.
- Zeng FQ, Xu HX, Sim KY, Gunsekera RM, Chen SX. 1998. The anticoagulant effects of Geum japonicum extract and its constituents. Phytother Res. 12(2):146–148.