Abstract
The complete chloroplast genome sequence of Pedicularis shansiensis Tsoong was determined and described. The circular chloroplast genome is 151,902 bp in length and contains a large single-copy region of 83,180 bp, a small single-copy region of 17,284 bp and two inverted repeat regions of 25,719 bp. The chloroplast genome contains 132 genes, including 86 protein-coding, 8 rRNA, and 38 tRNA genes. Phylogenetic analysis shows Pedicularis species cluster together and P. shansiensis forms a clade with P. dissect.
Pedicularis shansiensis Tsoong (1950) is perennial herb classified in the Orobanchaceae, which is an endemic species to China (Zhong Citation1963). It was reported that Pedicularis species showed anti-oxidation, anti-tumor, and anti-exercise fatigue activities (Chu et al. Citation2009; Zhu et al. Citation2012). Pedicularis is the largest hemiparasitic genus of Orobanchaceae. The plastome is more variable in heterotrophic plants (Li et al. Citation2021).
The fresh leaves were sampled from Zhuqueshan National Forest Park (33°79′N, 108°58′E; Shaanxi, China). The collection of material was in accordance with guidelines of China and Shaanxi Institute of International Trade and Commerce, and the voucher specimen (WQ202001) was deposited in the Shaanxi Institute of International Trade and Commerce Herbarium (https://yxy.csiic.com/, Wang Q, [email protected]). Total genomic DNA was extracted from leaves using the CTAB method, and sequenced on the Illumina HiSeq Sequencing System (Biomarker Technologies, Beijing).
The cleaned reads were assembled using GetOrganelle v1.7.2 using the default settings (Jin et al. Citation2020). The genome was annotated using CPGAVAS2 with 2544 published plastomes as references. The annotated chloroplast genome sequence was deposited in GenBank under the accession number MW542989.
The complete chloroplast genome of P. shansiensis is a circular DNA molecule 151,902 bp in length. The genome contains two inverted repeats (IRa and IRb; 25,719 bp each), one large single copy (LSC, 83,180 bp) and one small single copy (SSC, 17,284 bp). The overall GC content was 38.3% (LSC: 36.5%; SSC: 32.6%; IRs: 43.3%). The chloroplast genome contains 132 genes including 86 protein-coding, 8 rRNA, and 38 tRNA genes. The chloroplast genome of other Pedicularis species contained the 67–79 protein coding, 4 rRNA and 30 tRNA genes (Li et al. Citation2021).
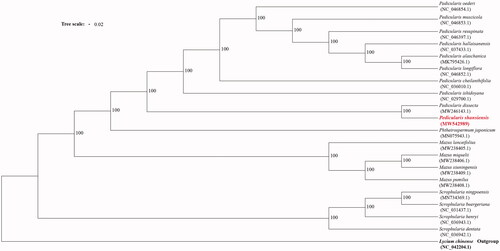
To investigate the phylogenetic position of P. shansiensis, a maximum likelihood tree was inferred using iqtree-2.1.2 (Lam-Tung et al. Citation2015) with the TVM + F + I + G4 model, and the bootstrap replicates were 1000 (). The results indicate that P. shansiensis was fully resolved in a clade with P. dissecta, and Pedicularis species cluster together, which is similar to previous research (Li et al. Citation2020).
Author contributions
Qi Wang: design and drafting of the paper; Zhipeng He, Ruirui Zhang and Xiaolin Dang: analysis; Xueyan Zhao: revising. All authors agree to be accountable for all aspects of the work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov/) under the accession no. MW542989. The associated BioProject, SRA, and BioSample numbers are PRJNA713966, SRS8526017, and SAMN18275831, respectively.
Additional information
Funding
References
- Chu HB, Tan NH, Peng CS. 2009. Progress in research on Pedicularis plants. Chin J Chin Mater Med. 34(19):2536–2546.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Lam-Tung N, Schmidt HA, Arndt VH, Quang BM. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Li X, Lin CY, Yang JB, Yu WB. 2020. De novo assembling a complete mitochondrial genome of Pedicularis rex (Orobanchaceae) using GetOrganelle toolkit. Mitochondrial DNA B Resour. 5(1):1056–1057.
- Li X, Yang JB, Wang H, Song Y, Corlett RT, Yao X, Li DZ, Yu WB. 2021. Plastid NDH pseudogenization and gene loss in a recently derived lineage from the largest hemiparasitic plant genus Pedicularis (Orobanchaceae). Plant Cell Physiol. 62(6):971–984.
- Zhong BQ. 1963. Flora of China. Vol. 68. Beijing: Science Press Ltd; p. 41.
- Zhu MJ, Cai CF, Tan NH, Xiong JY, Zhu HZ, Luo QJ. 2012. Anti-fatigue effects of extracts from Pedicularis densispica Franch on exercise mice. Nat Prod Res Dev. 24:1545–1548.